From Optflux
Revision as of 20:10, 26 February 2010 by Admin (talk | contribs) (Protected "EFM4OptFlux" [edit=sysop:move=sysop])
Contents
Example file
You can download it here -> Stelling_toy.xml (right-click + save link as...)
This tutorial uses this example from Z.Szallasi, J.Stelling and V.Periwal.System modeling in cell biology : from concepts to nuts and bolts. MIT Press, 2006
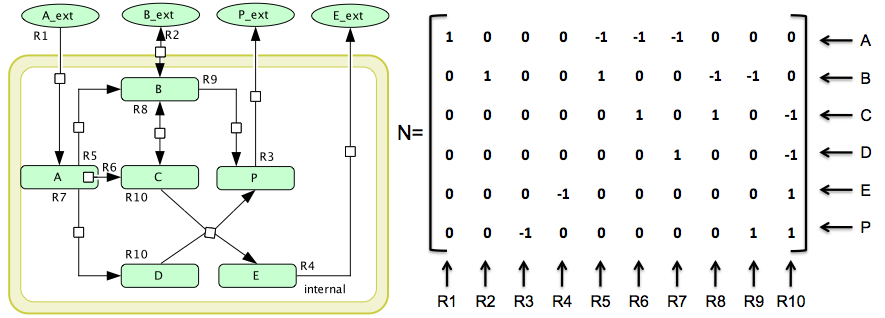
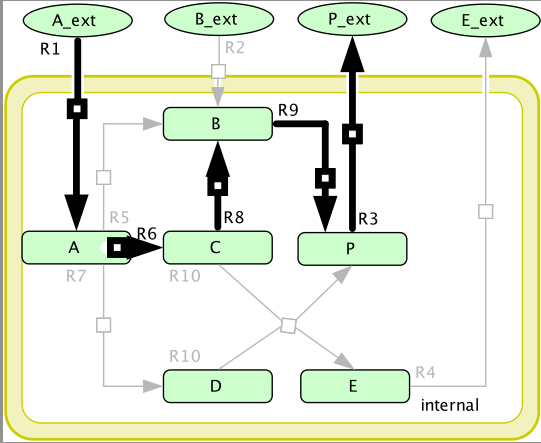
The simple network used in this example. N : q × m is the stoichiometric matrix where each row corresponds to one species and each column to one of the reactions. The reversibility information is given by rev = {R2, R8} and irrev = {R1, R3, R4, R5, R6, R7, R9, R10}, where rev is the set of reversible reactions and irrev is the set of irreversible ones. Also notice that rev ∩ irrev = ∅ .
Brief tutorial
This brief tutorial will help you to begin working with EFM4OptFlux.
- To start, you will need a valid CellDesigner SBML File loaded in OptFlux (see how to here). Remember to select CellDesigner SBML. You can use any model/format to calculate the EFMs, but in order to export to CellDesigner SBML, you will need a valid CellDesigner SBML file in the beginning.
Computing EFMs
- 1 - Start by accessing the menu Plugins->Elementary Modes->Compute Elementary Modes.
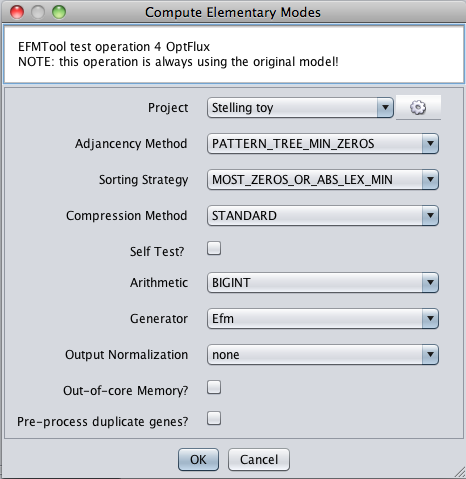
- 2 - The following dialog will be displayed. Arithmetics is constrained to BIGINT. We recommend using the default options, but feel free to explore...
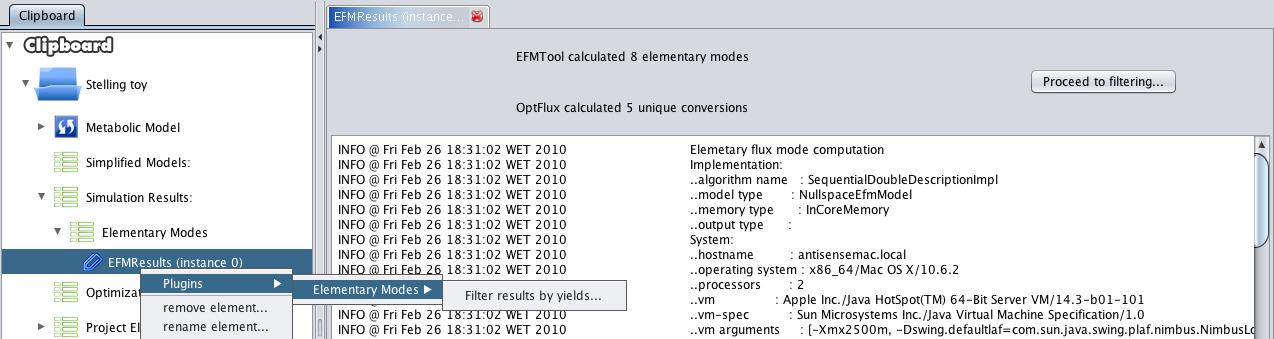
- 3 - After execution, a new datatype will be placed in the clipboard, under the simulation list -> elementary modes. Pressing it will launch a default viewer, summarizing the computation process.
Browsing EFMs using the filters
- 1 - Press proceed to filtering or use the context menu (depicted in the above screenshot).
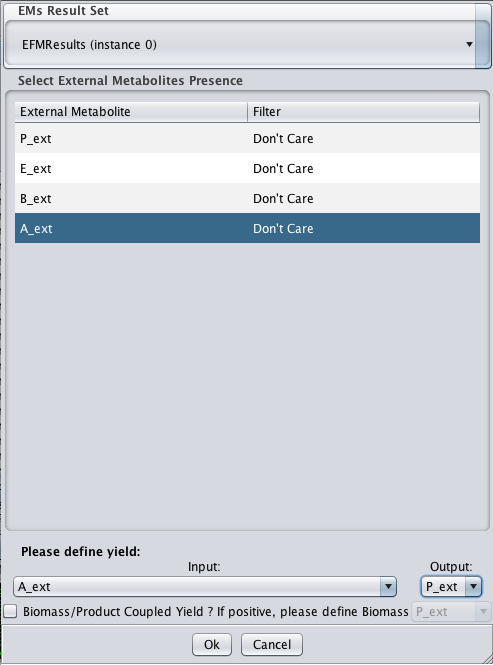
- 2 - You will be presented with the following dialog
- 3 - You should select which external metabolites you want to appear in the filtered conversions. The whole set of calculated net conversions will be filtered based on those choices. You should also select the desired yield. In the example all the filters are select to Don't Care, meaning that no filtering will actually be applied. The yield is defined as A_ext (Input) -> P_ext (Output). After the filtering step you should expect the following scenario.
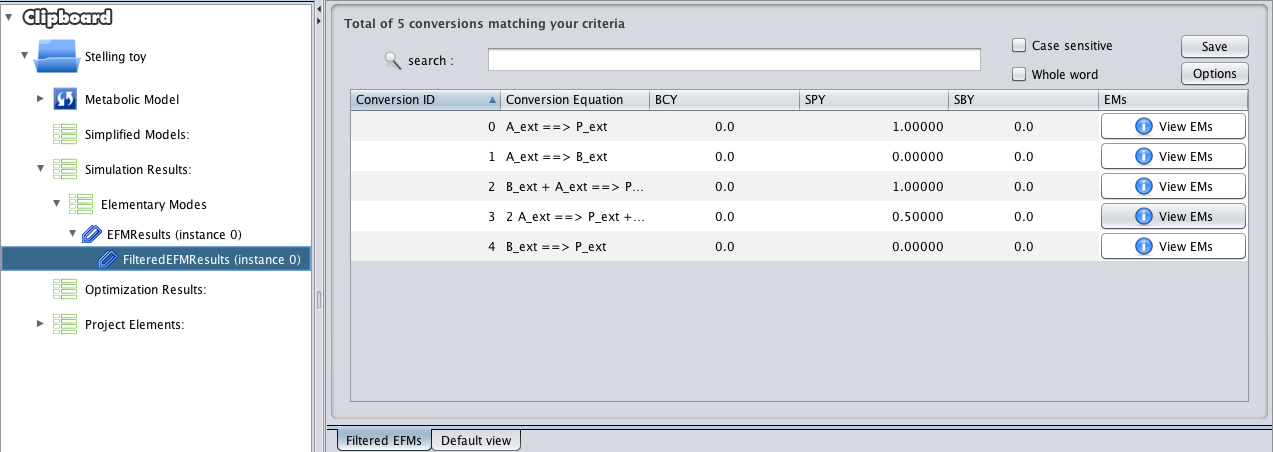
- 4 - In this panel you can browse the result of your filtering procedure. You can execute multiple filtring steps over the initial results, they will be added as a list on the bottom of those same original results (in the clipboard).
- This view allows sorting the conversions based on any column criteria (Conversion ID, Conversion Equation (Alphabetically), Biomass-Coupled Yield (BCY), Substrate-Product Yield (SPY) or Substrate-Biomass Yield (SBY)).
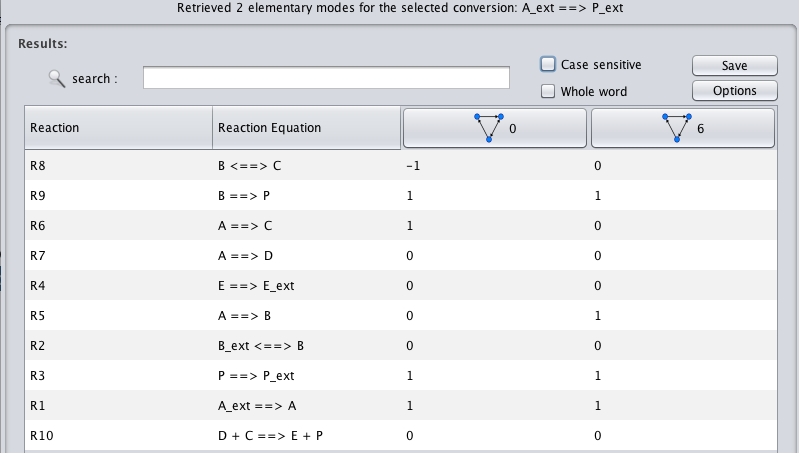
- By clicking the View EMs button for any conversion, the list of associated Elementary Modes will be displayed
- 5 - In this view, each row relates with a given reaction and each column relates with a given EM. By analyzing this column-wise table you can directly compare each EM for the reactions of your interest.
Exporting Results to CellDesigner
- 1 - In the above view, by clicking the header of each EM column you can export the EM relative values to a CellDesigner compatible file (assuming you loaded one in the beginning).
- 2 - Opening the resulting SBML file in CellDesigner will overlap the original CellDesigner layout with line widths reflecting the exported EM.