Simao.soares (talk | contribs) (→Import mappings) |
Simao.soares (talk | contribs) (→Import mappings) |
||
| Line 14: | Line 14: | ||
== Import mappings == | == Import mappings == | ||
| − | To import a mapping select the option MSeed -> Import -> Mappings and choose a name for the local data element and the local biochemistry to | + | To import a mapping select the option MSeed -> Import -> Mappings and choose a name for the local data element and the local biochemistry to be associated to the new mapping. OptFlux will then ask Model-seed to retrieve the mapping from The Seed and to create a local of version with the inserted name and connected to the selected local biochemistry.<br /> |
[[File:Import-mappings.png]] | [[File:Import-mappings.png]] | ||
Revision as of 16:29, 25 March 2013
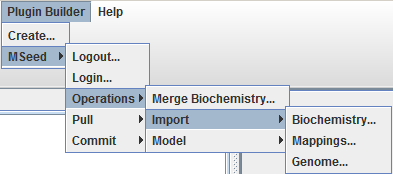
The Model-seed works in two levels, a local server and The seed server, it is possible to import to the local model-seed:
- biochemistry data
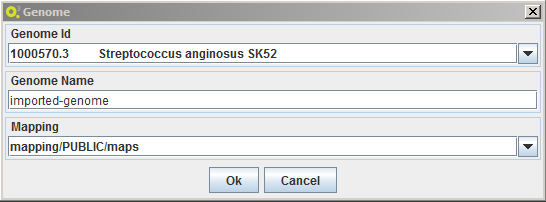
- genome annotations
- mappings
And use these data elements in local operations.
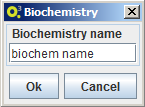
Import biochemistry
To import a biochemistry select the option MSeed -> Import -> Biochemistry and choose a name for the local data element. OptFlux will then ask Model-seed to retrieve the latest biochemistry version from The Seed and to create local of version with the selected name.
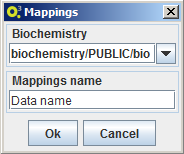
Import mappings
To import a mapping select the option MSeed -> Import -> Mappings and choose a name for the local data element and the local biochemistry to be associated to the new mapping. OptFlux will then ask Model-seed to retrieve the mapping from The Seed and to create a local of version with the inserted name and connected to the selected local biochemistry.