(→How to import database reactions) |
|||
| Line 38: | Line 38: | ||
The mutant simulation with addiction of reactions can be access in “Plugins -> Add Reactions->Simulation” menu. | The mutant simulation with addiction of reactions can be access in “Plugins -> Add Reactions->Simulation” menu. | ||
<br> | <br> | ||
| − | + | [[Image:AddReactionsSimulation.png]] | |
<br><br><b><i>Select knockouts and added reactions</i></b><br> | <br><br><b><i>Select knockouts and added reactions</i></b><br> | ||
After choose the database, you can select a set of reactions from “Reactions” list using the buttons (arrows). The reactions that will be added to the model appear in the “Add Reactions” box. The same process is used to select the list of knockouts. | After choose the database, you can select a set of reactions from “Reactions” list using the buttons (arrows). The reactions that will be added to the model appear in the “Add Reactions” box. The same process is used to select the list of knockouts. | ||
Revision as of 17:24, 9 December 2011
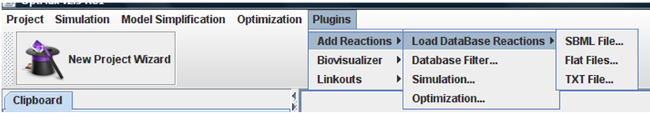
All features of plug-in can be accessed under the “Plugins-> Add Reactions” menu.
Contents
How to import database reactions
You can import database by three options:
1. SBML file
Choose the option “SBML File” in menu. It will be appear a new window when you can choose the project, SBML file and the name of database.
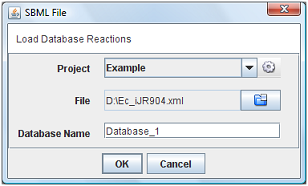
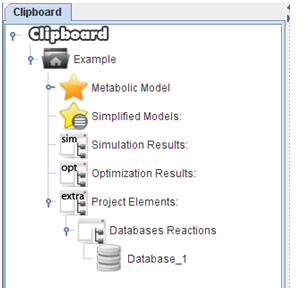
After press “Ok”, the all the reactions will be loaded into the clipboard.
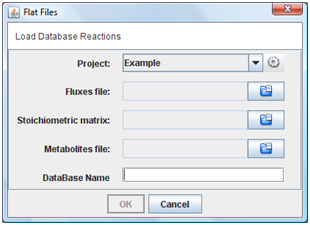
2. Flat files
If you choose “Flat Files” option in menu, you must indicate the metabolites, fluxes and stoichiometric matrix text files.
3. TXT Files
This is a new format representation for reactions and metabolites. In this case, you must indicate the metabolites and reactions text files. More information about files format can be found here.
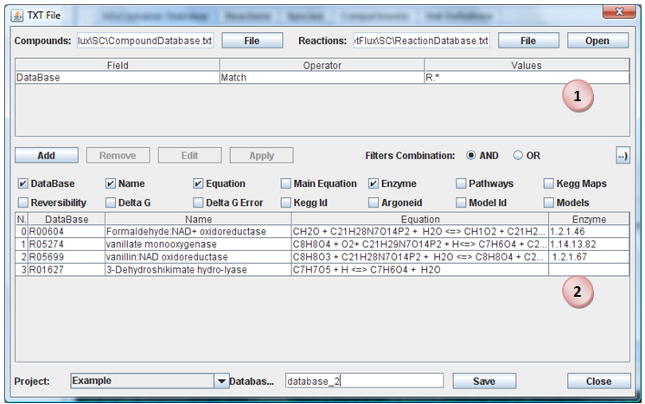
Choose Files
Select the reactions and metabolites files. Click in “Open” button.
Apply Filters
You can select, by applying filters, a subset of reactions. For do this, click in “Add” to insert a new filter. When you have all filters that you want apply, click in “Apply” button. The filters can be applied in conjunction or disjunction of them. You can remove or edit a filter by select it and click in button “Remove” or “Edit”.
Import Reactions
When all reactions that you want import are present in reactions table, you need select the Project, a name for database and press the “Save” button. A new database will be created in selected project with the reactions visible in table (2).
How to reduce the database of reactions
To reduce the number of reactions in database to optimization process, you can apply a filter giving the target compound and a value of tolerance. The process will found all pathways between the metabolites presented in model and the target compound and calculate the cost for each pathway. Using the tolerance value will be selected the pathways with less cost and others that are in tolerance range.
This process can be accessed in "AddReactions->Database Filter".
How to do a mutant simulation by adding reactions
The mutant simulation with addiction of reactions can be access in “Plugins -> Add Reactions->Simulation” menu.
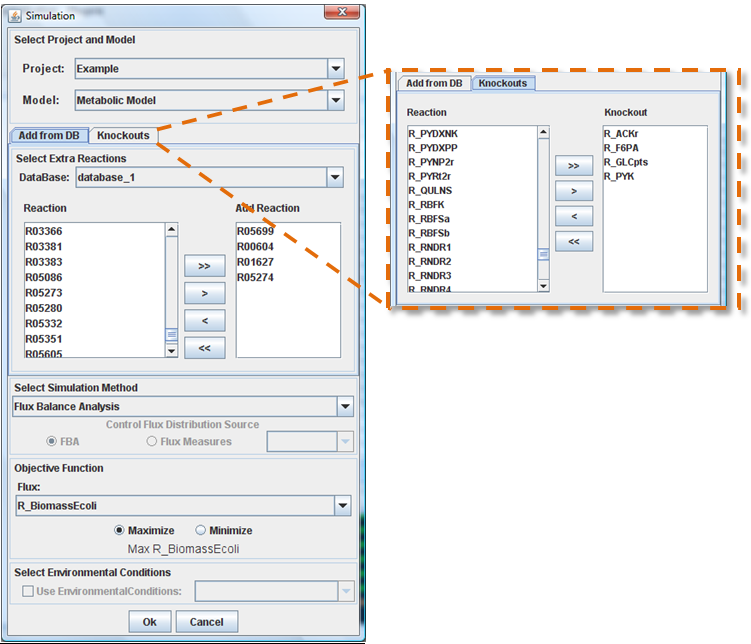
Select knockouts and added reactions
After choose the database, you can select a set of reactions from “Reactions” list using the buttons (arrows). The reactions that will be added to the model appear in the “Add Reactions” box. The same process is used to select the list of knockouts.
Select Simulation Method
OptFlux can use several simulation methods, namely: Flux Balance Analysis, ROOM-LP, ROOM-MILP and MOMA. When you only add reactions to the model, only the FBA method should be used.
ROOM-LP stands for the Regulatory On-Off Minimization Method (ROOM), using a linear programming (LP) relaxation; ROOM-MILP is the original ROOM that uses a Mixed Integer LP (MILP) method; MOMA stands for the Minimization of Metabolic Adjustment method that uses quadratic programming.
Objective Function Configuration
Here you can select the reaction to optimize (biomass, by default), and you can also define if you will be maximizing or minimizing the flux.
Select Environmental Conditions
If you have created environmental conditions you can select them to be used as constrains in the simulation. These can be used to define the values of drain fluxes, i.e. the rates at which metabolites are consumed or produced.
Finally, you can press Ok button and the results will be loaded into the clipboard.