| (5 intermediate revisions by the same user not shown) | |||
| Line 9: | Line 9: | ||
<br> | <br> | ||
http://sites.google.com/site/optfluxhowtos/howtos/1/newProjectWizard.png | http://sites.google.com/site/optfluxhowtos/howtos/1/newProjectWizard.png | ||
| − | <br> | + | <br><br> |
<b>1st step</b><br> | <b>1st step</b><br> | ||
In the 1st step of the wizard you have to enter a name to the project and choose SBML as the model source input.<br> | In the 1st step of the wizard you have to enter a name to the project and choose SBML as the model source input.<br> | ||
| − | |||
<div style="display: block; text-align: left;"> | <div style="display: block; text-align: left;"> | ||
| − | http://sites.google.com/site/optfluxhowtos/howtos/1/Step1NewProjectSBML.png</div> | + | http://sites.google.com/site/optfluxhowtos/howtos/1/Step1NewProjectSBML.png</div><br> |
| − | </b><br> | + | <b>2nd step</b><br> |
OptFlux | OptFlux | ||
| − | allows you to load | + | allows you to load Standard SBML files and also CellDesigner SBML files.<br> |
In | In | ||
the 2nd step, you have to define the type of the file, and also choose | the 2nd step, you have to define the type of the file, and also choose | ||
| Line 28: | Line 27: | ||
<b> | <b> | ||
[[Image:Step2 sbml.png]] | [[Image:Step2 sbml.png]] | ||
| − | </b><br> | + | </b><br><br> |
<b>3th step</b><br> | <b>3th step</b><br> | ||
| − | + | To allow OptFlux to determine the external metabolites you can choose to use "Use compartments heuristics" or "Use regular expressions". | |
| + | <br> | ||
| + | The first detects external metabolites based on the definitions of the compartments contained in the SBML file. | ||
| + | Using the regular expressions option you can easily set up an expression to detect external metabolites from patterns in their names using the options: "starting with" / "ending with" / "contains" / "match regexp".<br> | ||
| + | You can also load the most commonly used regular expressions using the option "Load common regexps". | ||
| + | <br> | ||
<br> | <br> | ||
[[Image:Step3 sbml.png]] | [[Image:Step3 sbml.png]] | ||
| Line 38: | Line 42: | ||
The identification of the reaction corresponding to biomass formation is an essential task for performing simulations. In this step you have to validate the biomass/ growth reaction automatically detected. You can change it if the heuristic used in the detection did not work.<br> | The identification of the reaction corresponding to biomass formation is an essential task for performing simulations. In this step you have to validate the biomass/ growth reaction automatically detected. You can change it if the heuristic used in the detection did not work.<br> | ||
<br> | <br> | ||
| − | [[Image:Step4 | + | [[Image:Step4.png]] |
<br> | <br> | ||
<br> | <br> | ||
Latest revision as of 17:39, 18 November 2009
How to create a new model/project from SBML
In OptFlux, metabolic models are the basis for all the work. The concept of Project gathers all data generated for a given model. Therefore, projects can be created providing a model. In this How To, we show how to create a model, given an SBML file with the required information. To know more about the SBML format check http://www.sbml.org.
Start the
creation of a new project using the New Project Wizard. You can access
it either through the Project menu or the Toolbar.
http://sites.google.com/site/optfluxhowtos/howtos/1/newProjectWizard.png
1st step
In the 1st step of the wizard you have to enter a name to the project and choose SBML as the model source input.
2nd step
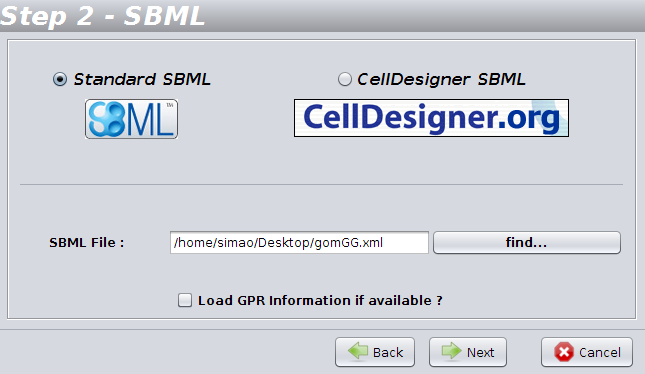
OptFlux
allows you to load Standard SBML files and also CellDesigner SBML files.
In
the 2nd step, you have to define the type of the file, and also choose
the name and location of the file you want to load.
If you want to load GPR
(gene-protein-reaction) information from the SBML file, choose the option
"Load GPR information if available".
3th step
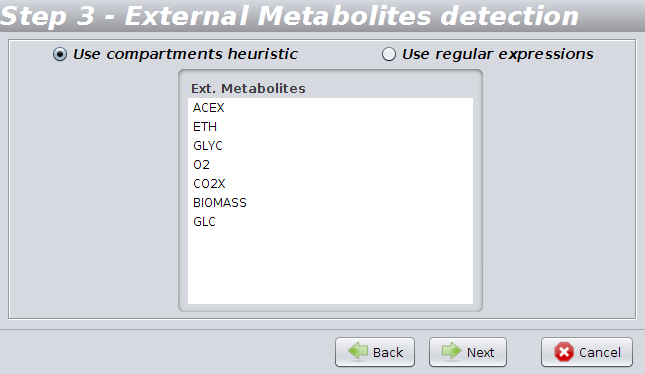
To allow OptFlux to determine the external metabolites you can choose to use "Use compartments heuristics" or "Use regular expressions".
The first detects external metabolites based on the definitions of the compartments contained in the SBML file.
Using the regular expressions option you can easily set up an expression to detect external metabolites from patterns in their names using the options: "starting with" / "ending with" / "contains" / "match regexp".
You can also load the most commonly used regular expressions using the option "Load common regexps".
4th step
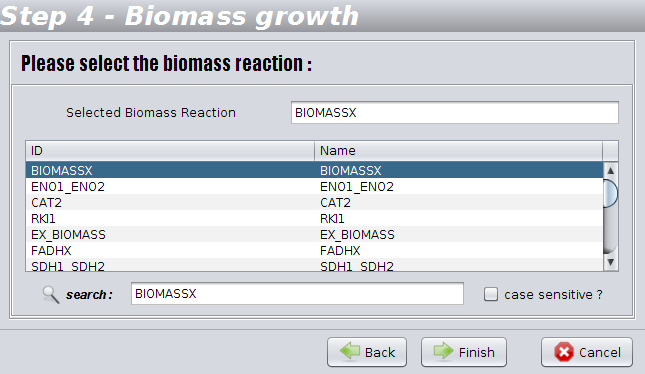
The identification of the reaction corresponding to biomass formation is an essential task for performing simulations. In this step you have to validate the biomass/ growth reaction automatically detected. You can change it if the heuristic used in the detection did not work.
And that's all! After created, the new Project will be available in the clipboard and you can enjoy the OptFlux features.