| (2 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | {{DISPLAYTITLE: Knockout simulations}} | ||
OptFlux allows the user to perform a simulation of a mutant strain, i.e. of the model with some genetic modifications, in this case a set of gene or reaction deletions. | OptFlux allows the user to perform a simulation of a mutant strain, i.e. of the model with some genetic modifications, in this case a set of gene or reaction deletions. | ||
| Line 26: | Line 27: | ||
<br> | <br> | ||
<b>4.Select the simulation method</b><br> | <b>4.Select the simulation method</b><br> | ||
| − | Select | + | Select between the two simulation methods available for wild type: FBA and pFBA<br><br> |
<i><b>4.Select Environmental Conditions</b></i><br> | <i><b>4.Select Environmental Conditions</b></i><br> | ||
If you have created environmental conditions you can select them to be used as constraints in the optimization. These can be used to define the values of drain fluxes, i.e. the rates at which metabolites are consumed or produced.<br> | If you have created environmental conditions you can select them to be used as constraints in the optimization. These can be used to define the values of drain fluxes, i.e. the rates at which metabolites are consumed or produced.<br> | ||
Latest revision as of 18:43, 12 December 2012
OptFlux allows the user to perform a simulation of a mutant strain, i.e. of the model with some genetic modifications, in this case a set of gene or reaction deletions.
Access the "Knockout" option either through the "Simulation" menu or right clicking on the model icon in the clipboard.
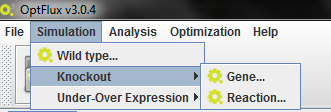
In this menu you wil have two options: "Gene" and "Reaction".
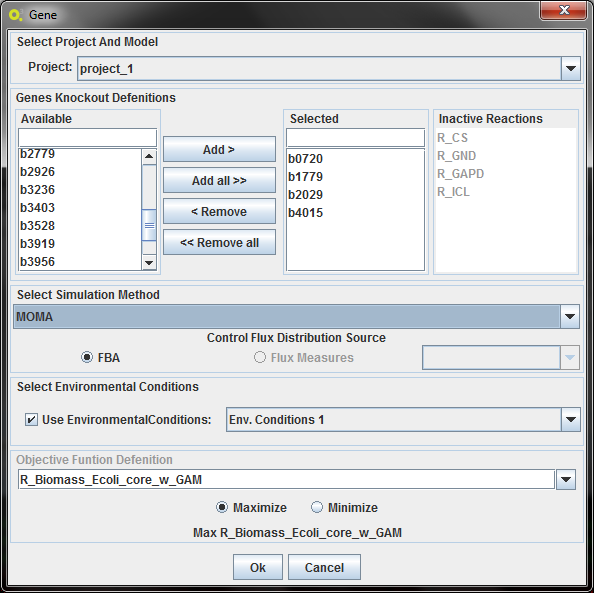
By selecting "Gene" you will have a interface that allows you to select the genes you want to knockout/delete, and the reactions associated with that gene which will be knocked out/deleted because of that.
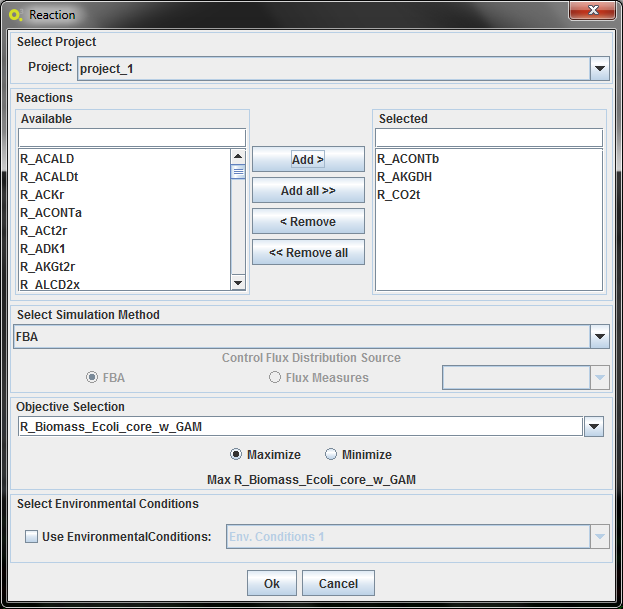
Selecting Reaction will allow the selection of the reactions directly.
You can simulate a mutant in OptFlux followign the steps:
1.Select Project
Choose the Project to do the simulation.
2.Select Genes/Reactions to delete
Choose the gene/reactions as shown above.
3.Objective Function Configuration
In the objective function section, you have select a reaction (typically the biomass/ growth reaction, selected by default) to maximize or minimize the corresponding flux.
4.Select the simulation method
Select between the two simulation methods available for wild type: FBA and pFBA
4.Select Environmental Conditions
If you have created environmental conditions you can select them to be used as constraints in the optimization. These can be used to define the values of drain fluxes, i.e. the rates at which metabolites are consumed or produced.
And that's all!! You can press OK and the results will be loaded into the clipboard.