(→Exporting networks) |
(→Note about graphical visualizing) |
||
| (11 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
TNA4OptFlux networks are created by extracting data from the mathematical models used by OptFlux to run simulations, in OptFlux each mathematical model is associated with a project consequently before using TNA4OptFlux at least one project has to exist, created networks are associated with the project from whose modal they derived. | TNA4OptFlux networks are created by extracting data from the mathematical models used by OptFlux to run simulations, in OptFlux each mathematical model is associated with a project consequently before using TNA4OptFlux at least one project has to exist, created networks are associated with the project from whose modal they derived. | ||
| − | |||
| − | |||
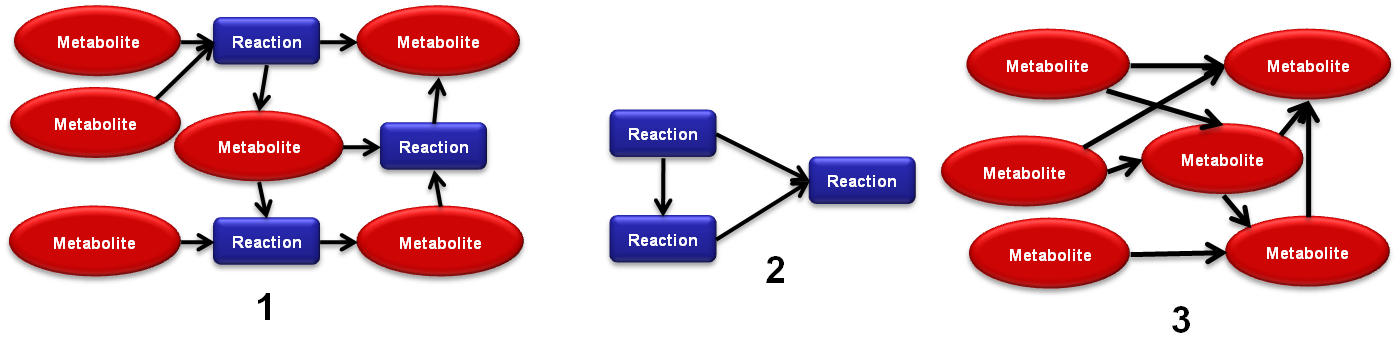
[[Image:TNA4OptFlux1.png|frame|right|700x176px|1) Reaction-Metabolite network; 2) Reaction only networks; 3) Metabolite only network]] | [[Image:TNA4OptFlux1.png|frame|right|700x176px|1) Reaction-Metabolite network; 2) Reaction only networks; 3) Metabolite only network]] | ||
| − | + | <br> | |
To create a network: | To create a network: | ||
| Line 25: | Line 23: | ||
* 3 - HITS (Hubs-and-authorities) | * 3 - HITS (Hubs-and-authorities) | ||
* 4 - Clustering coefficient | * 4 - Clustering coefficient | ||
| − | + | <br> | |
To use any ranking algorithm just go to '''Analysis -> TNA4 -> Ranking algorithms''' and select the desired one from the menu. | To use any ranking algorithm just go to '''Analysis -> TNA4 -> Ranking algorithms''' and select the desired one from the menu. | ||
| Line 35: | Line 33: | ||
* 2 - MBNF format - this is the default format of InBiNA, the network analysis aplication that TNA4OptFlux is based on. | * 2 - MBNF format - this is the default format of InBiNA, the network analysis aplication that TNA4OptFlux is based on. | ||
* 3 - XGMML format - this is an XML format which cytoscape supports, it is especially useful when dealing with variation networks because the flux values are also exported. | * 3 - XGMML format - this is an XML format which cytoscape supports, it is especially useful when dealing with variation networks because the flux values are also exported. | ||
| − | + | <br> | |
To export a network go to '''Analysis -> TNA4 -> Export''' and select the desired format. | To export a network go to '''Analysis -> TNA4 -> Export''' and select the desired format. | ||
= Shortest path metrics = | = Shortest path metrics = | ||
| − | + | Besides simply identifying the shortest paths between two vertices, TNA4OptFlux supports several other analysis functionalities base in the shortest path. | |
| − | |||
| − | |||
| − | + | To use TNA4OptFlux's shortest path functionalities : | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | * 1 - Select to '''Analysis -> TNA4 -> Get Shortest Path Metrics'''. | |
| + | * 2 - Select the network. | ||
| + | * 3 - There is now a "shortest path metrics" object associated with the network which contains the shortest path information divided by four panels: | ||
| + | ** 3.1 - The first panels contains the generic networks shortest path data: the diameter, average shortest path, larger shortest path in the network and number of vertices with inward and outward edges. | ||
| + | ** 3.2 - The second panel allows user to calculate the distance between two select vertices alternatively the user can visualize one shortest path or all possible shortest path. | ||
| + | ** 3.3 - The third panel can be used to obtain generic information about individual vertices. | ||
| + | ** 3.4 - Finally the fourth panel contains a table with an histogram of the shortest path. | ||
| − | + | = Independent modules (or subgraphs) = | |
| − | To | + | |
| + | Some networks contain subgraphs, or independent modules, these are parts of the network which are isolated from the rest of the network, independent modules shouldn't be present in metabolic networks obtained directly from a model, but when dealing with filtered networks they can occur | ||
| + | <br> | ||
| + | To identify the independent modules present in a network using TNA4OptFlux select '''Analysis -> TNA4 -> Indentify independent modules''', the independent modules can then be converted into networks if more in-depth analysis of them is required. | ||
= Filters = | = Filters = | ||
| − | Filters | + | Filters are used to create new networks by removing elements from existing ones which can be useful when dealing with extensive networks or when only part of one network is of interest. |
| − | To use | + | |
| − | + | * 1 - To use a filter go to '''Analysis -> TNA4 -> Filter network'''. | |
| + | * 2 - Select the network, the project it is associated with and the name of the filtered network to be created. | ||
| + | * 3 - Chose if the filtered network should contain drain reactions and/or external metabolites. | ||
| + | * 4 - If there are simulations associated with the project decide if simulation filtering (removal of the parts of the network which were not active or preformed under a selected threshold in a simulated scenario) should be used. | ||
| + | * 5 - Chose the type of degree based filtering to be executed (if any), currently the types supported are: | ||
| + | ** 5.1 - Removal of metabolites with a inward degree above a set threshold. | ||
| + | ** 5.2 - Removal of metabolites with a outward degree above a set threshold. | ||
| + | ** 5.3 - Removal of metabolites with a degree above a set threshold. | ||
| + | ** 5.4 - Removal of the k metabolites (value chosen by user) with higher inward degree. | ||
| + | ** 5.5 - Removal of the k metabolites (value chosen by user) with higher outward degree. | ||
| + | ** 5.6 - Removal of the k metabolites (value chosen by user) with higher degree. | ||
| + | * 6 - If any ranking algorithm was used decide if it should be used as an filtering metric. | ||
| + | * 7 - Select individual vertices to remove. | ||
| + | * 8 - Select individual edges to remove. | ||
<br> | <br> | ||
| + | It should be noted that regardless of the filtering method choose vertices with an degree of zero will be removed from the final network. | ||
| + | |||
| + | = Degree distribution = | ||
| + | |||
| + | To calculate the degree distribution of a network select '''Analysis -> TNA4 -> Degree distribution'''. | ||
| + | |||
| + | = Network comparison = | ||
| + | |||
| + | Using TNA4OptFlux it is possible to compare a pair of networks to do so select '''Analysis -> TNA4 -> Compare networks''' and select the network pair. Comparisons are made using the network structure and any analyses metrics which were applied to booth of the networks previously to the comparison. | ||
<br> | <br> | ||
| − | + | Most of the comparison functionalities are straightforward however one deserves further explanation: the concept of decision points, these are metabolites which are present in booth networks but are consumed by different sets of reactions each. Decision points received their name because they can be considered parts of the network were there was a "decision" about the flux matter in the metabolism. | |
| + | |||
| + | = Locating active vertices = | ||
| + | |||
| + | The location of active vertices is an functionality of TNA4OptFlux which can be used to give an idea of which reactions should active if certain metabolites are present in the environment. It is a recursive algorithm based in the petri net like structure that a Reaction-Metabolite networks posses. To use this functionality: | ||
| + | * 1 - Go to '''Analysis -> TNA4 -> Locate Active Vertices'''. | ||
| + | * 2 - Select from the list of metabolites the ones which are supposed to be present in the environment. | ||
<br> | <br> | ||
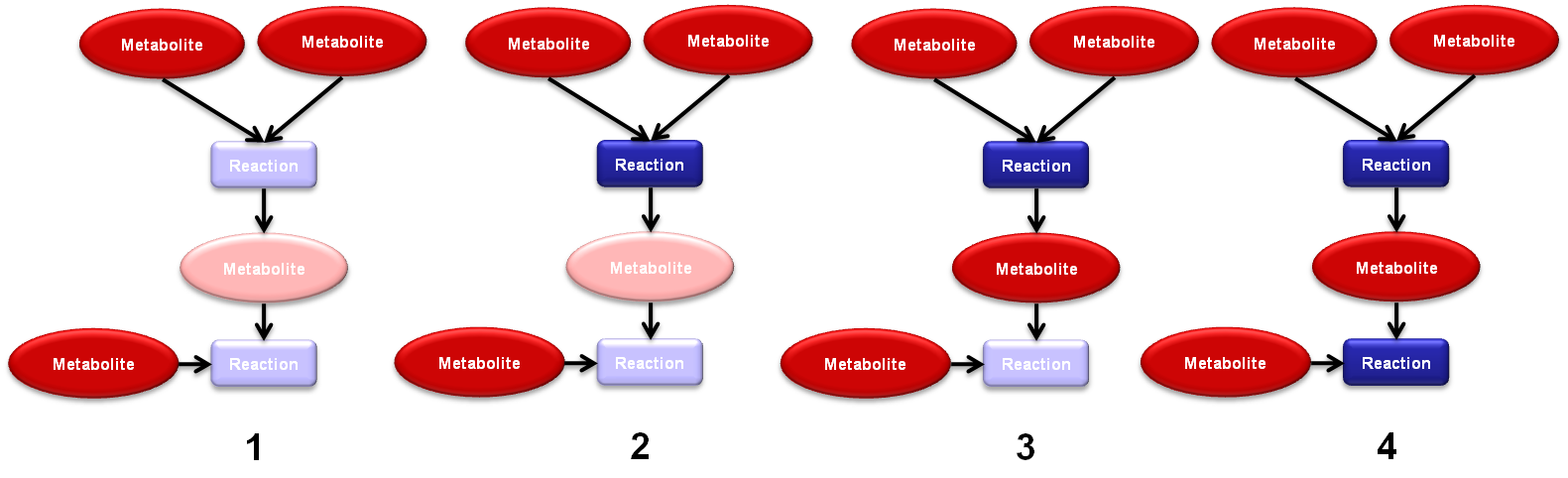
| + | [[Image:TNA4OptFlux2.png|frame|center|790x241px|Example of the location of active vertices algorithm]] | ||
<br> | <br> | ||
| − | + | Please not that this functionality only works with Reaction-Metabolite networks. | |
| − | + | ||
| − | + | = Calculating flux variation = | |
| − | + | ||
| − | + | When simulation filtering is used the resulting subnetwork contains the values of the reactions fluxes which were used in the filtering operation. TNA4OptFlux can be used to compare the flux values associated with an networks obtained via simulation filtering, this functionality doesn't pertain directly to network analysis but it can help users compeering different simulation which complements the analysis of variation networks nicely. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<br> | <br> | ||
| + | To use compare the flux values: | ||
| + | * 1 - Go to '''Analysis -> TNA4 -> Calculate flux variation'''. | ||
| + | * 2 - Chose the project and network. | ||
| + | * 3 - Select an reference network, alternatively the fluxes can be compared with the results of an wildtype simulation obtained via pFBA. | ||
<br> | <br> | ||
| + | Please note that only networks that were created though simulation filtering have associated flux values, consequently they are the only ones that can be used with this analysis tool. | ||
| + | |||
| + | = Variation networks = | ||
| − | + | The concept of variation networks was development by combining network comparison with simulation filtering, essentially these are networks which are obtained by comparing two or more networks derived from the same base using different filters and creating a third network with only the parts which differed from one network to another (the variations). This variation network can then be used as a tool to compare the different simulated situations which can be particularly useful for the analysis of a mutant organism if it is compared with a wildtype. | |
<br> | <br> | ||
| + | Initially the concept of variation networks worked only with a pair of networks but latter it was expanded to work with solution sets, in this case one network is taken as reference (usually the wildtype) and it is compared with each network in the solution se, in the end the variation network obtained contains only the more commune variations, based in an user defined threshold. | ||
<br> | <br> | ||
| − | + | TNA4OpfFlux uses two variation metrics: exclusivity (a vertex is exclusive if it belongs only to one network) and flux variation (if a reaction flux changed over a user defined threshol). | |
<br> | <br> | ||
| + | To create an variation network from a pair o networks with associated flux data: | ||
| + | * 1 - Go to Analysis -> TNA4 -> Varition networks -> create variation network from network pair. | ||
| + | * 2 - Select the two networks, booth must belong to the same project and have associated flux data. | ||
| + | * 3 - Select the variation metrics to use. | ||
| + | ** 3.1 - If flux variation is used a threshold, either absolute value or an percentage (0.0 to 1.0) must be selected. | ||
<br> | <br> | ||
| − | * 4 - | + | To create an variation network from a solution set: |
| + | * 1 - Go to Analysis -> TNA4 -> Varition networks -> create variation network from solution set. | ||
| + | * 2 - Select a project and a wild type simulation result. | ||
| + | * 3 - Select the simulation results to be compared to the wild type. | ||
| + | *4 - Select the variation metrics to use and the minimal threshold. | ||
| + | **4.1 - If flux variation is used a threshold, either absolute value or an percentage (0.0 to 1.0) must be selected | ||
<br> | <br> | ||
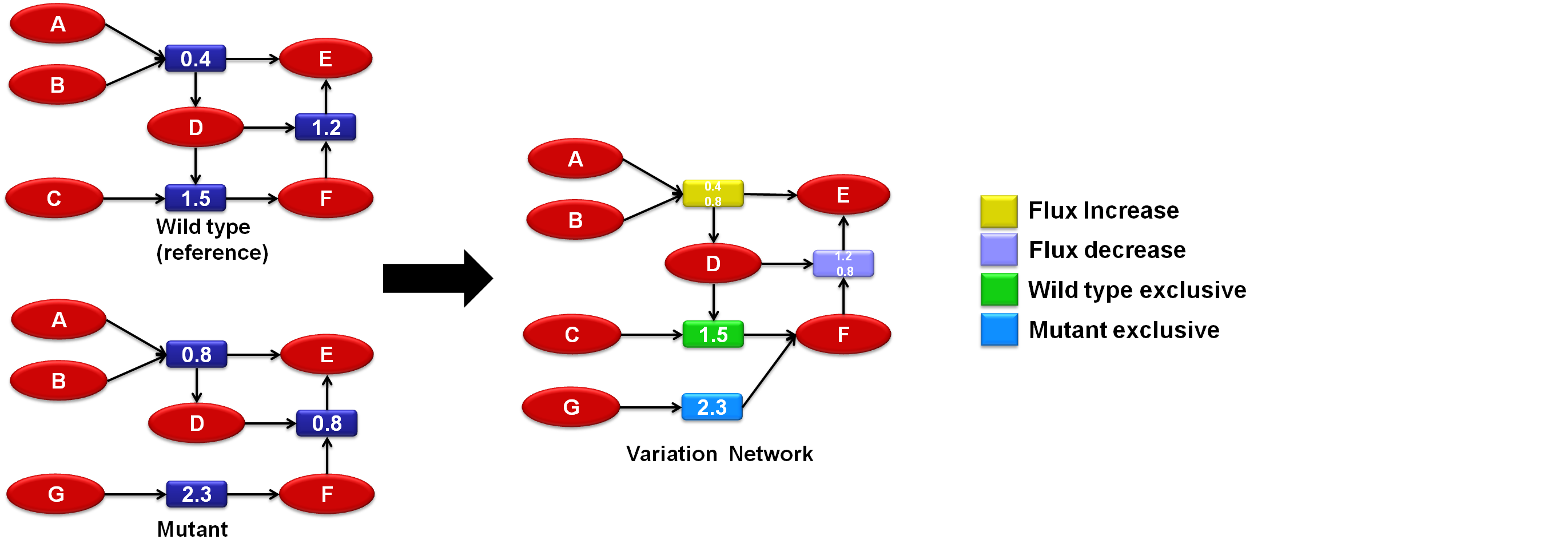
| + | [[Image:TNA4OptFlux3.png|frame|center|914x319px|Sample variation network]] | ||
<br> | <br> | ||
| − | + | Please note that when creating a variation network from a solution set it is not necessary to first create a network for each solution. | |
| − | |||
| − | |||
| − | |||
| − | + | = Note about graphical visualizing = | |
| − | + | [[Image:TNA4OptFlux4.png|frame|right|611x175px|Vertex colors used in TNA4OptFlux graphical functionalities]] | |
| − | = | + | Because of their large size it is impossible for most part to fully draw a metabolic network, TNA4OptFlux is capable however of representing graphically small subsections of a network. |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<br> | <br> | ||
| + | The graphical representation functionality of TNA4OptFlux draws circular graphs centered around a selected vertices, this graphs contain the central vertex and the vertices connected to it though a path with a length of up to five edges. It is possible to navigate the network though the graphical representation by click in a vertex doing so marks it as the central vertex and redraws the graph. | ||
<br> | <br> | ||
| − | + | The graph can be moved by dragging and dropping with left mouse bottom and zoom in and out is achieved through the use of the right one. By default blue vertices represent metabolites and yellow ones reactions however when dealing with variation networks different colors are used to identify exclusive reactions. | |
<br> | <br> | ||
| − | + | [[Image:TNA4OptFlux5.png|frame|left|511x298px|TNA4OptFlux graphical visualization]] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 17:19, 15 January 2013
Contents
- 1 How to create a network
- 2 Using ranking algorithms
- 3 Exporting networks
- 4 Shortest path metrics
- 5 Independent modules (or subgraphs)
- 6 Filters
- 7 Degree distribution
- 8 Network comparison
- 9 Locating active vertices
- 10 Calculating flux variation
- 11 Variation networks
- 12 Note about graphical visualizing
How to create a network[edit]
TNA4OptFlux networks are created by extracting data from the mathematical models used by OptFlux to run simulations, in OptFlux each mathematical model is associated with a project consequently before using TNA4OptFlux at least one project has to exist, created networks are associated with the project from whose modal they derived.
To create a network:
- 1 - Go to Analysis -> TNA4 -> New network.
- 2 - Select the project, the type of the network, name your new network and decide if it should include drain and/or external reactions. There are three possible types of networks:
- 2.1 - Reaction-Metabolite networks: these are bipartite graphs where the vertices represent the reactions and the metabolites.
- 2.2 - Metabolite only networks: only contain metabolite vertices and the edges connect the compounds which are converted from one to another by the reactions.
- 2.3 - Reaction only networks only contain reaction vertices and the edges connect reactions to the ones which consume their products (Warning: the creating of this networks usually takes significantly more time than the other two types).
- 3 - If filters are used then rest of the process is identical to apply a filter to an already existing network.
Using ranking algorithms[edit]
Ranking algorithm was classification given to any algorithm which analysis a network and give to each vertex a ranking value, following this definition TNA4OptFlux implements four ranking algorithm:
- 1 - Betweenness Centrality
- 2 - Closeness Centrality
- 3 - HITS (Hubs-and-authorities)
- 4 - Clustering coefficient
To use any ranking algorithm just go to Analysis -> TNA4 -> Ranking algorithms and select the desired one from the menu.
Exporting networks[edit]
TNA4OptFlux networks can be exported as files, currently three file formats are suported:
- 1 - Pajek format - this format is used by the Pajek network analysis application, networks exported in this format are converted to two files: a network file and a partition file.
- 2 - MBNF format - this is the default format of InBiNA, the network analysis aplication that TNA4OptFlux is based on.
- 3 - XGMML format - this is an XML format which cytoscape supports, it is especially useful when dealing with variation networks because the flux values are also exported.
To export a network go to Analysis -> TNA4 -> Export and select the desired format.
Shortest path metrics[edit]
Besides simply identifying the shortest paths between two vertices, TNA4OptFlux supports several other analysis functionalities base in the shortest path.
To use TNA4OptFlux's shortest path functionalities :
- 1 - Select to Analysis -> TNA4 -> Get Shortest Path Metrics.
- 2 - Select the network.
- 3 - There is now a "shortest path metrics" object associated with the network which contains the shortest path information divided by four panels:
- 3.1 - The first panels contains the generic networks shortest path data: the diameter, average shortest path, larger shortest path in the network and number of vertices with inward and outward edges.
- 3.2 - The second panel allows user to calculate the distance between two select vertices alternatively the user can visualize one shortest path or all possible shortest path.
- 3.3 - The third panel can be used to obtain generic information about individual vertices.
- 3.4 - Finally the fourth panel contains a table with an histogram of the shortest path.
Independent modules (or subgraphs)[edit]
Some networks contain subgraphs, or independent modules, these are parts of the network which are isolated from the rest of the network, independent modules shouldn't be present in metabolic networks obtained directly from a model, but when dealing with filtered networks they can occur
To identify the independent modules present in a network using TNA4OptFlux select Analysis -> TNA4 -> Indentify independent modules, the independent modules can then be converted into networks if more in-depth analysis of them is required.
Filters[edit]
Filters are used to create new networks by removing elements from existing ones which can be useful when dealing with extensive networks or when only part of one network is of interest.
- 1 - To use a filter go to Analysis -> TNA4 -> Filter network.
- 2 - Select the network, the project it is associated with and the name of the filtered network to be created.
- 3 - Chose if the filtered network should contain drain reactions and/or external metabolites.
- 4 - If there are simulations associated with the project decide if simulation filtering (removal of the parts of the network which were not active or preformed under a selected threshold in a simulated scenario) should be used.
- 5 - Chose the type of degree based filtering to be executed (if any), currently the types supported are:
- 5.1 - Removal of metabolites with a inward degree above a set threshold.
- 5.2 - Removal of metabolites with a outward degree above a set threshold.
- 5.3 - Removal of metabolites with a degree above a set threshold.
- 5.4 - Removal of the k metabolites (value chosen by user) with higher inward degree.
- 5.5 - Removal of the k metabolites (value chosen by user) with higher outward degree.
- 5.6 - Removal of the k metabolites (value chosen by user) with higher degree.
- 6 - If any ranking algorithm was used decide if it should be used as an filtering metric.
- 7 - Select individual vertices to remove.
- 8 - Select individual edges to remove.
It should be noted that regardless of the filtering method choose vertices with an degree of zero will be removed from the final network.
Degree distribution[edit]
To calculate the degree distribution of a network select Analysis -> TNA4 -> Degree distribution.
Network comparison[edit]
Using TNA4OptFlux it is possible to compare a pair of networks to do so select Analysis -> TNA4 -> Compare networks and select the network pair. Comparisons are made using the network structure and any analyses metrics which were applied to booth of the networks previously to the comparison.
Most of the comparison functionalities are straightforward however one deserves further explanation: the concept of decision points, these are metabolites which are present in booth networks but are consumed by different sets of reactions each. Decision points received their name because they can be considered parts of the network were there was a "decision" about the flux matter in the metabolism.
Locating active vertices[edit]
The location of active vertices is an functionality of TNA4OptFlux which can be used to give an idea of which reactions should active if certain metabolites are present in the environment. It is a recursive algorithm based in the petri net like structure that a Reaction-Metabolite networks posses. To use this functionality:
- 1 - Go to Analysis -> TNA4 -> Locate Active Vertices.
- 2 - Select from the list of metabolites the ones which are supposed to be present in the environment.
Please not that this functionality only works with Reaction-Metabolite networks.
Calculating flux variation[edit]
When simulation filtering is used the resulting subnetwork contains the values of the reactions fluxes which were used in the filtering operation. TNA4OptFlux can be used to compare the flux values associated with an networks obtained via simulation filtering, this functionality doesn't pertain directly to network analysis but it can help users compeering different simulation which complements the analysis of variation networks nicely.
To use compare the flux values:
- 1 - Go to Analysis -> TNA4 -> Calculate flux variation.
- 2 - Chose the project and network.
- 3 - Select an reference network, alternatively the fluxes can be compared with the results of an wildtype simulation obtained via pFBA.
Please note that only networks that were created though simulation filtering have associated flux values, consequently they are the only ones that can be used with this analysis tool.
Variation networks[edit]
The concept of variation networks was development by combining network comparison with simulation filtering, essentially these are networks which are obtained by comparing two or more networks derived from the same base using different filters and creating a third network with only the parts which differed from one network to another (the variations). This variation network can then be used as a tool to compare the different simulated situations which can be particularly useful for the analysis of a mutant organism if it is compared with a wildtype.
Initially the concept of variation networks worked only with a pair of networks but latter it was expanded to work with solution sets, in this case one network is taken as reference (usually the wildtype) and it is compared with each network in the solution se, in the end the variation network obtained contains only the more commune variations, based in an user defined threshold.
TNA4OpfFlux uses two variation metrics: exclusivity (a vertex is exclusive if it belongs only to one network) and flux variation (if a reaction flux changed over a user defined threshol).
To create an variation network from a pair o networks with associated flux data:
- 1 - Go to Analysis -> TNA4 -> Varition networks -> create variation network from network pair.
- 2 - Select the two networks, booth must belong to the same project and have associated flux data.
- 3 - Select the variation metrics to use.
- 3.1 - If flux variation is used a threshold, either absolute value or an percentage (0.0 to 1.0) must be selected.
To create an variation network from a solution set:
- 1 - Go to Analysis -> TNA4 -> Varition networks -> create variation network from solution set.
- 2 - Select a project and a wild type simulation result.
- 3 - Select the simulation results to be compared to the wild type.
- 4 - Select the variation metrics to use and the minimal threshold.
- 4.1 - If flux variation is used a threshold, either absolute value or an percentage (0.0 to 1.0) must be selected
Please note that when creating a variation network from a solution set it is not necessary to first create a network for each solution.
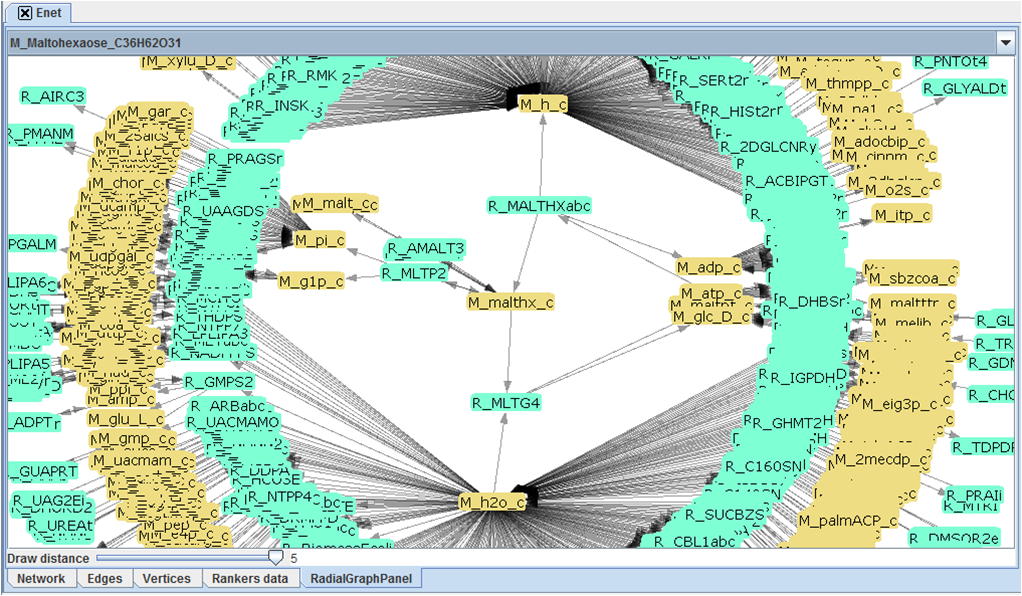
Note about graphical visualizing[edit]
Because of their large size it is impossible for most part to fully draw a metabolic network, TNA4OptFlux is capable however of representing graphically small subsections of a network.
The graphical representation functionality of TNA4OptFlux draws circular graphs centered around a selected vertices, this graphs contain the central vertex and the vertices connected to it though a path with a length of up to five edges. It is possible to navigate the network though the graphical representation by click in a vertex doing so marks it as the central vertex and redraws the graph.
The graph can be moved by dragging and dropping with left mouse bottom and zoom in and out is achieved through the use of the right one. By default blue vertices represent metabolites and yellow ones reactions however when dealing with variation networks different colors are used to identify exclusive reactions.