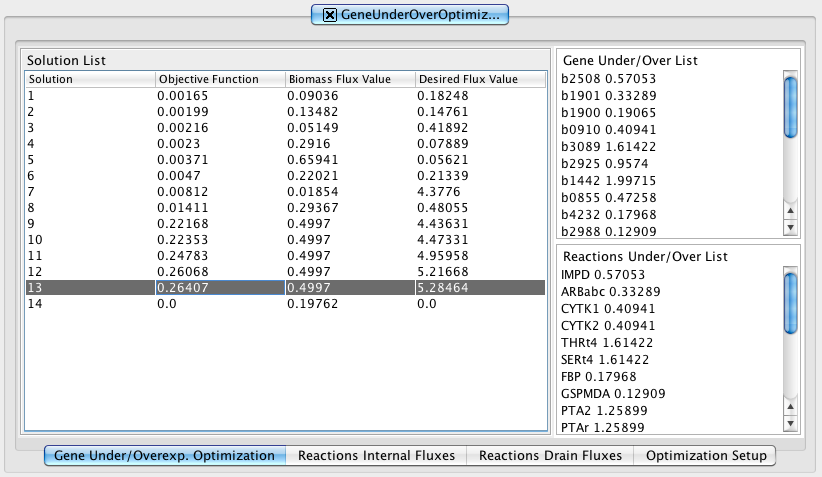
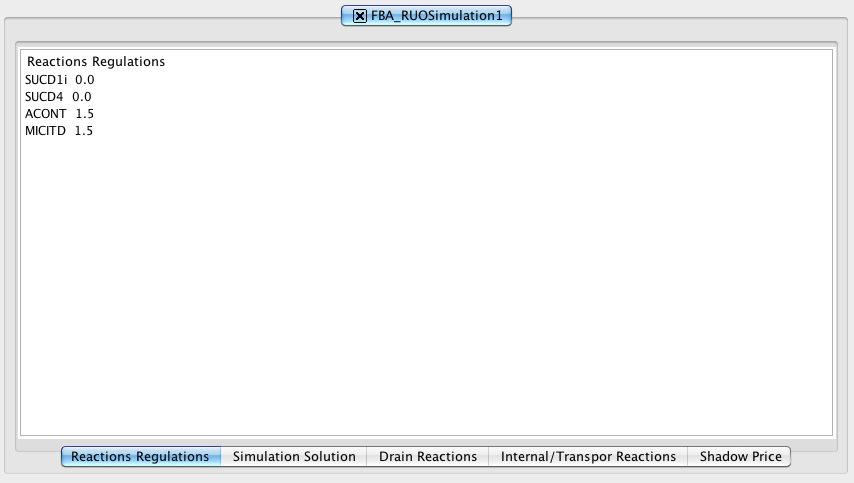
1. How to do a mutant simulation - Reactions Regulations
You can access the under/overexpression reaction simulation option under the “Plugins -> Under and Overexpression -> Reaction Simulation…”.
In the Reaction Simulation you can select the model/project to work, and set up your configuration.
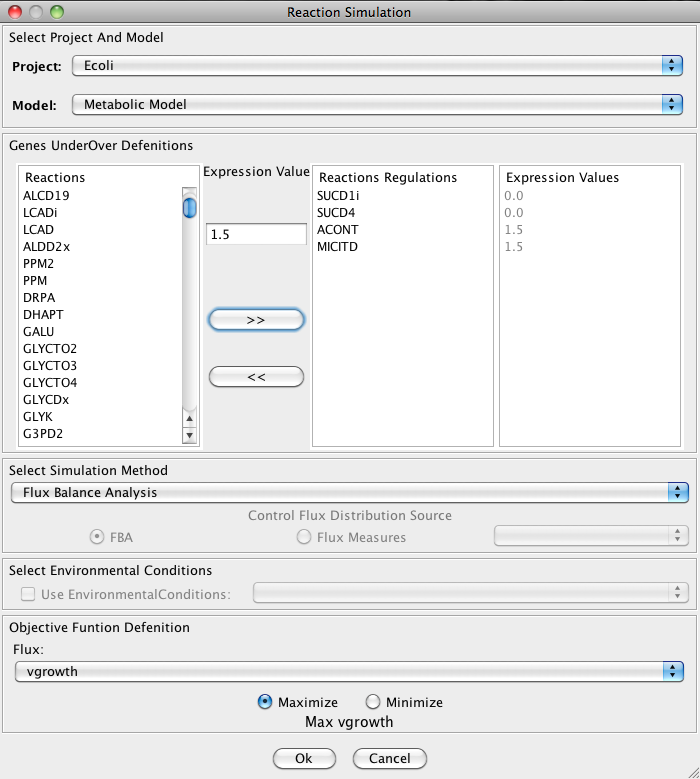
1. Reaction Regulations Set Up
Selecting in the Reactions list and defining an expression value in Expression Value box you can add/remove (using the arrows buttons) reactions regulations. On the right side appear the Reactions Regulations list and the respective Expression Value.
2. Select Simulation Method
OptFlux can use several simulation methods for knockout simulations, namely: Flux-Balance Analysis, ROOM-LP, ROOM-MILP, MOMA
ROOM-LP stands for the Regulatory On-Off Minimization Method (ROOM), using a linear programming (LP) relaxation; ROOM-MILP is the original ROOM that uses a Mixed Integer LP (MILP) method; MOMA stands for the Minimization of Metabolic Adjustment method that uses quadratic programming.
3. Objective Function Configuration
Here you can select the reaction to optimize (biomass, by default), and you can also define if you will be maximizing or minimizing the flux.
4. Select Environmental Conditions
If you have created environmental conditions you can select them to be used as constrains in the simulation. These can be used to define the values of drain fluxes, i.e. the rates at which metabolites are consumed or produced.
Finally, you can press Ok button and the results will be loaded into the clipboard.
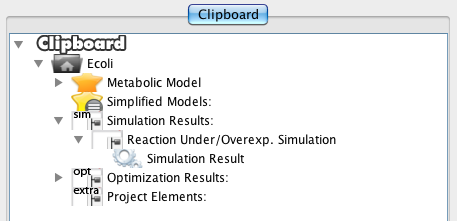
Double-click on the Simulation Result will show the results panel.
2. How to do a mutant simulation - Genes Regulations
You can access the under/overexpression reaction simulation option under the “Plugins -> Under and Overexpression -> Gene Simulation…”.

In the Gene Simulation you can select the model/project to work, and set up your configuration.
File:GeneSimulationMenu.png.png
1. Gene Regulations Set Up
Selecting in the Genes list and defining an expression value in Expression Value box you can add/remove (using the arrows buttons) reactions regulations. On the right side appear the Genes Regulations list and the respective Expression Value, and also the influenced reactions regulations.
2. Select Simulation Method
OptFlux can use several simulation methods for knockout simulations, namely: Flux-Balance Analysis, ROOM-LP, ROOM-MILP, MOMA
ROOM-LP stands for the Regulatory On-Off Minimization Method (ROOM), using a linear programming (LP) relaxation; ROOM-MILP is the original ROOM that uses a Mixed Integer LP (MILP) method; MOMA stands for the Minimization of Metabolic Adjustment method that uses quadratic programming.
3. Objective Function Configuration
Here you can select the reaction to optimize (biomass, by default), and you can also define if you will be maximizing or minimizing the flux.
4. Select Environmental Conditions
If you have created environmental conditions you can select them to be used as constrains in the simulation. These can be used to define the values of drain fluxes, i.e. the rates at which metabolites are consumed or produced.
Finally, you can press Ok button and the results will be loaded into the clipboard.
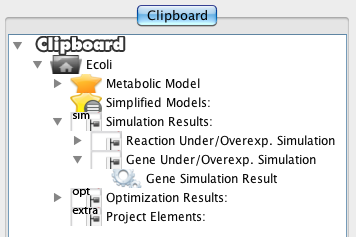
Double-click on the Gene Simulation Result will show the results panel.