| Line 34: | Line 34: | ||
*1 - After loaded the metabolic model in OptFlux, accessing the '''Load Regulatory Net''' option under the '''Plugins->Regulatory Too''' menu | *1 - After loaded the metabolic model in OptFlux, accessing the '''Load Regulatory Net''' option under the '''Plugins->Regulatory Too''' menu | ||
[[Image:PathForIntegratedModel.png]] | [[Image:PathForIntegratedModel.png]] | ||
| + | |||
*2 - The following dialog will be displayed. | *2 - The following dialog will be displayed. | ||
| Line 46: | Line 47: | ||
<b>2. <i>File Options</i></b> | <b>2. <i>File Options</i></b> | ||
| − | You can select the regulatory model file | + | You can select the regulatory model file. In our example the regulatory model file name is '''toy.regNet'''. |
| + | |||
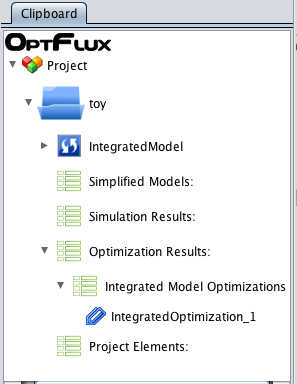
*3 - After execution, the '''Metabolic Model''' datatype will be replaced for '''Integrated Model''' in the clipboard. | *3 - After execution, the '''Metabolic Model''' datatype will be replaced for '''Integrated Model''' in the clipboard. | ||
[[Image:ClipBoardAfterIntegrateModel.png]] | [[Image:ClipBoardAfterIntegrateModel.png]] | ||
| + | ==How to do a mutant simulation== | ||
| − | |||
[[Image:PathForSimAndOpt.png]] | [[Image:PathForSimAndOpt.png]] | ||
Revision as of 09:07, 9 July 2010
Contents
Example file
You can download it here ->
This tutorial uses this example based on an example from J. Kim and J. Reed.OptORF: Optimal metabolic and regulatory perturbations for metabolic engineering of microbial strains. BMC, 2010
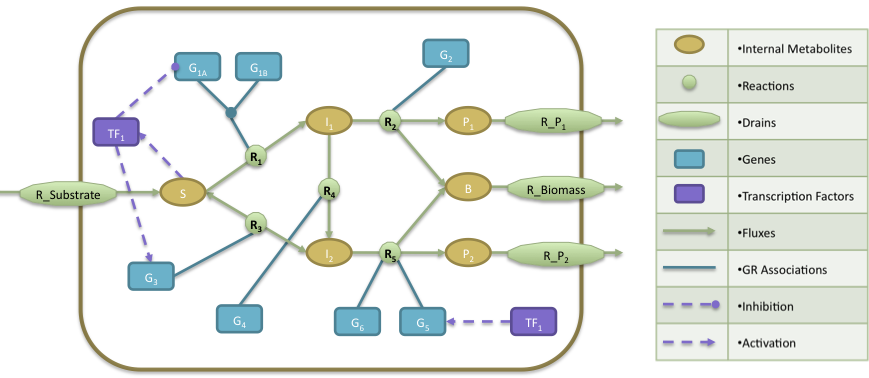
- The regulatory model is composed by a transcription factor (TF1) that is activated when metabolite S is present, activates the expression of two genes (G3, G5) and represses the expression of gene G1A.
- The Gene Reactions rules said us that: R1 is catalyzed when the gene G1A and G1B are activated; R2 is catalyzed by the gene G2; R3 and R4 are catalyzed by the genes G3 and G4, respectively; finally, R5 can be catalyzed by either genes G5 or G6.
- In the metabolic level, the substrate (S) is utilized to produce biomass (B) and by-products P1 and P2. The cellular objective is to maximize biomass production (B) and the engineering objective is the production of P1. Reaction R2 converts the internal metabolite I1 into product P1 and 0.08 biomass (B), whereas reaction R5 converts the internal metabolite I2 into product P2 and 0.12 Biomass. The stoichiometric coefficients of all other reactions reflect a one-to-one relationship between molecule quantities.
Brief tutorial
This brief tutorial will help you to begin working with RegulatoryTool4OptFlux.
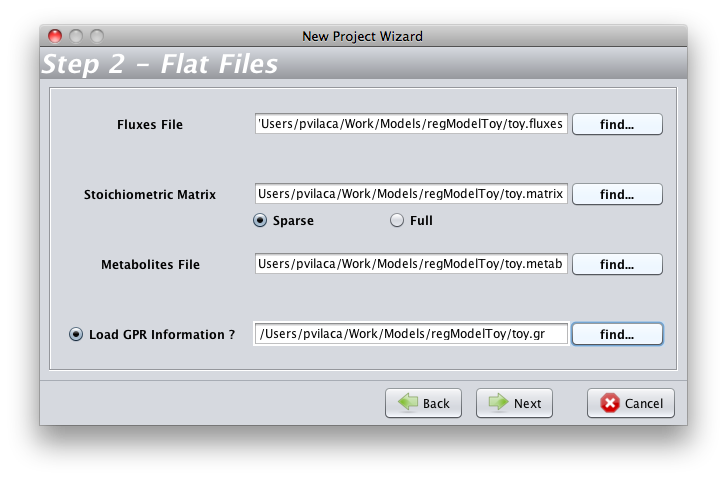
- To start, you will need load a model in OptFlux(see how to here). In the New Project Wizard you have to chose the the toy.fluxes file as the Fluxes File; the toy.matrix file as the Stoichiometric Matrix; the toy.metab file as the Metabolites File and the toy.gr file as the Load GPR Information.
- On the wizard Step3, you have to chose indexing started at zero(0)
- You must pay attention to select a correct biomass flux (R_Biomass).
How to integrate a regulatory model
To start using all features present in Regulatory plug-in, you have to integrate a regulatory model with a metabolic model previous loaded in OptFlux.
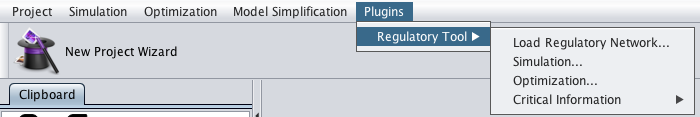
- 1 - After loaded the metabolic model in OptFlux, accessing the Load Regulatory Net option under the Plugins->Regulatory Too menu
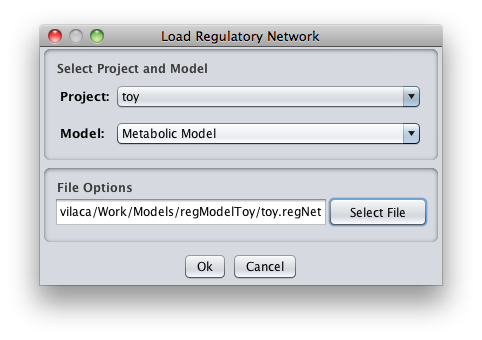
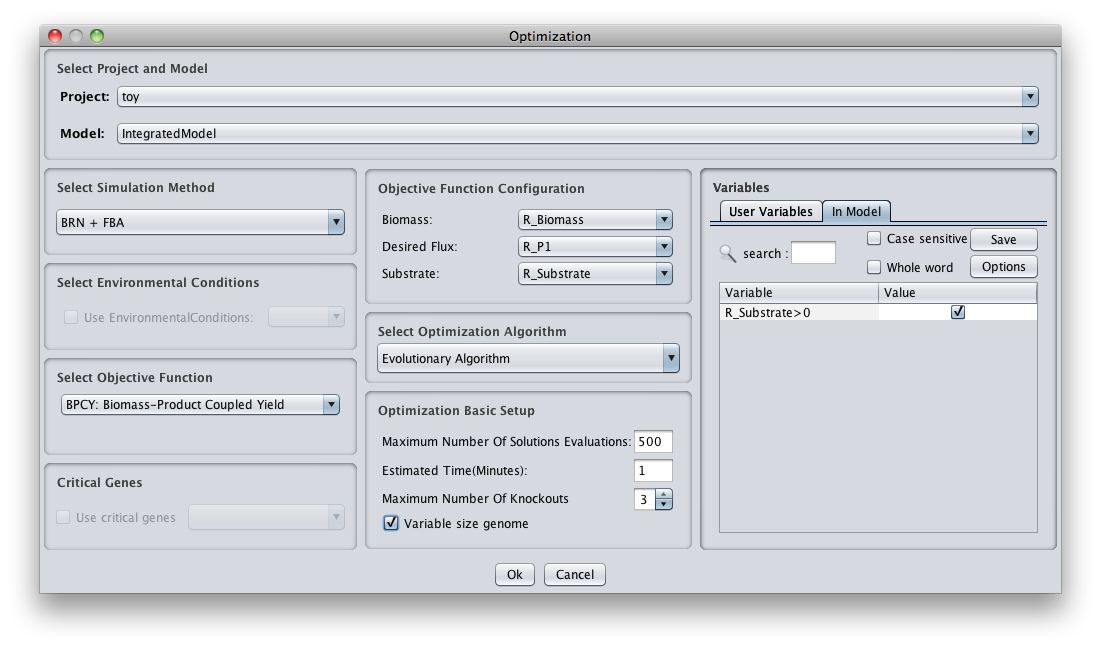
- 2 - The following dialog will be displayed.
1. Select Project and Model
You can select the model/project to integrate the regulatory model.
2. File Options
You can select the regulatory model file. In our example the regulatory model file name is toy.regNet.
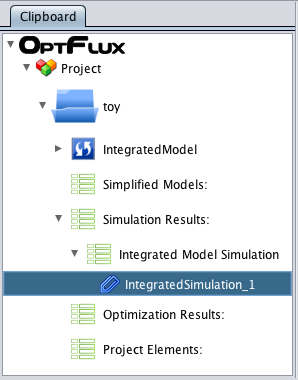
- 3 - After execution, the Metabolic Model datatype will be replaced for Integrated Model in the clipboard.
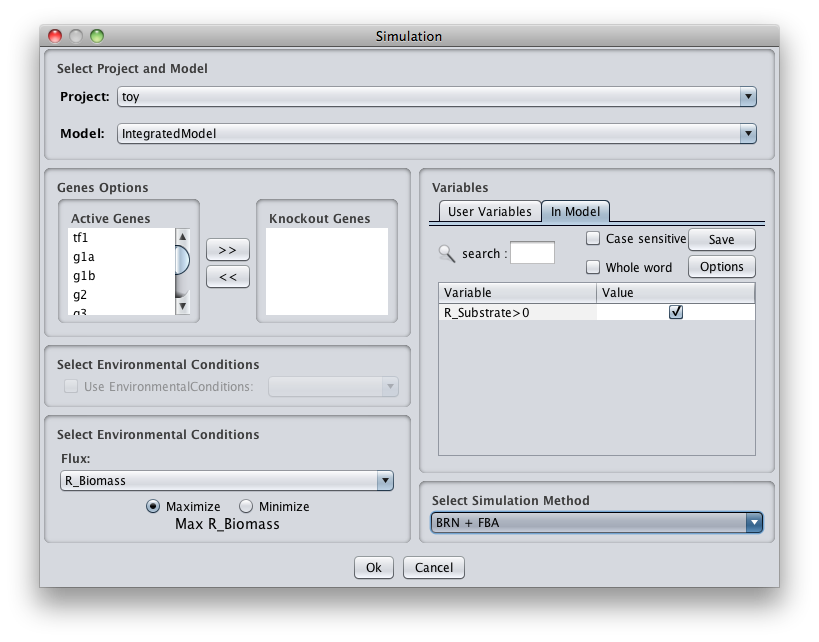
How to do a mutant simulation
Critical Genes
The regulatory plug-in allows the users to define which are the genes present on the integrated model that are essential for the strain to survive, i.e. to keep a value of growth (biomass reaction flux) different from zero. The essential genes can be calculated using BRN+FBA method. Also, the software allows the user to load these genes or reactions from a file (if they are available). The list of essential genes can be manually edited, allowing users to add or remove elements, given their knowledge or the purpose of their experiments.
How to do load critical genes from a file
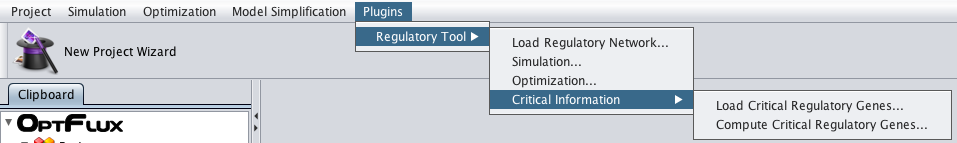
- To load the information from a file just access the "Load Critical Regulatory Genes" option under the "Plugins->Regulatory Tool->Critical Information" menu in the Project Menu or right click on the Integrated Model icon in the clipboard.
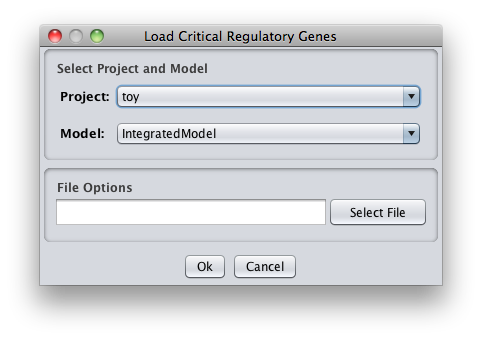
- Choose the name of the project and the model to where you want to load the critical information and also the file that have the critical genes.
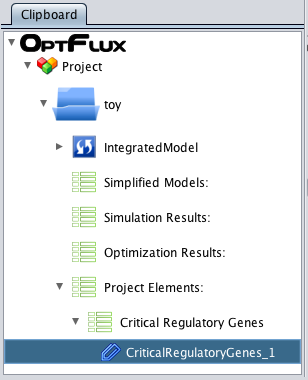
- Now press OK and you can see a new element on the clipboard with the loaded critical genes.
How to determining essential genes
- You can access the "Compute Critical Regulatory Genes" option under the "Plugins->Regulatory Tool->Critical Information" menu or by right clicking on the Integrated Model icon on the clipboard.
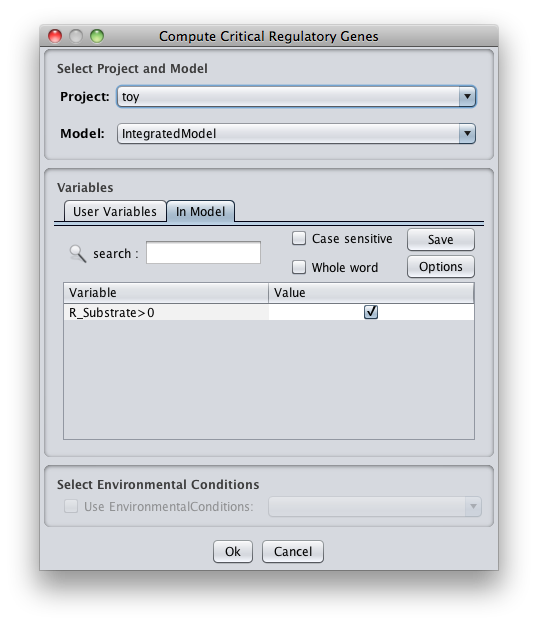
- In the options you can select the model/project to work, and set up the process.
1.Set Regulatory User Variables
Choose the value of all regulatory user variables.
2. Select Environmental Conditions
If you have created environmental conditions you can select them to be used as constraints in the critical genes/reactions calculation. This means that in this case you will calculate essential genes in a given environment.
- And that's all !! Now you can press OK and check the results in the clipboard.