These import layout operations are availabe in the Import menu:
There are 2 major operations for layout importation in OptFlux. The default import operation allows importing layouts from XGMML, COBRA, CellDesigner and SBGN layouts.
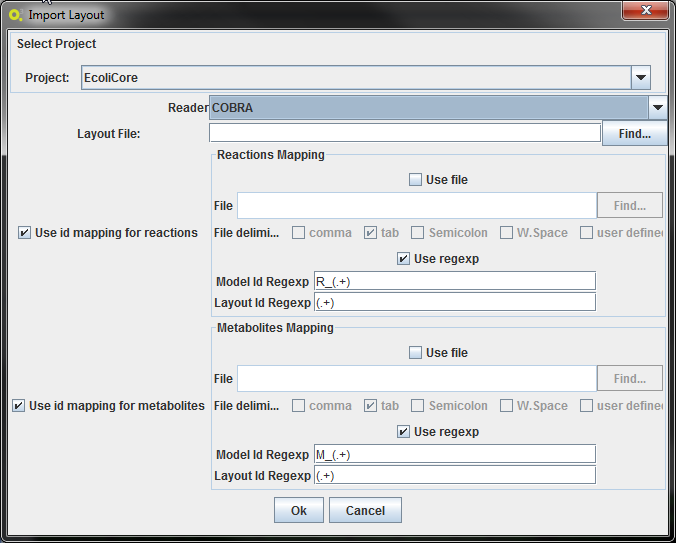
In this interface it is possible to define the mapping of the id's from the metabolic model to the layout. This mapping may not be necessary, but this way it is possible, for instance, to use the same layout for different models.
The mapping can be made two ways:
- Providing a 2-column file: mapping the metabolic model identifier to the layout identifier
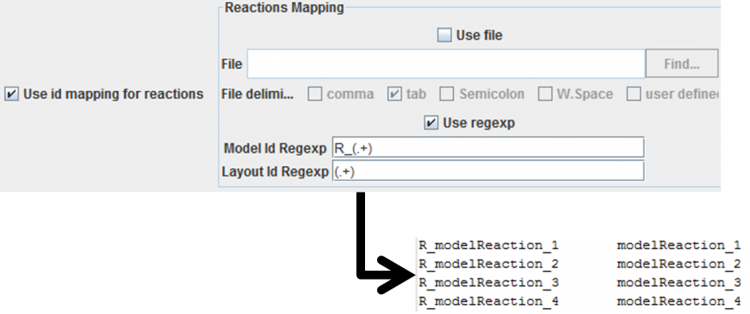
- Applying regexps to the ids: the regexps must contain only one group. If the id's match, by applying those regexp's, then that layout entity will be mapped with the metabolic model entity. An example of a reaction mapping can be seen below.
Importing a KGML Layout[edit]
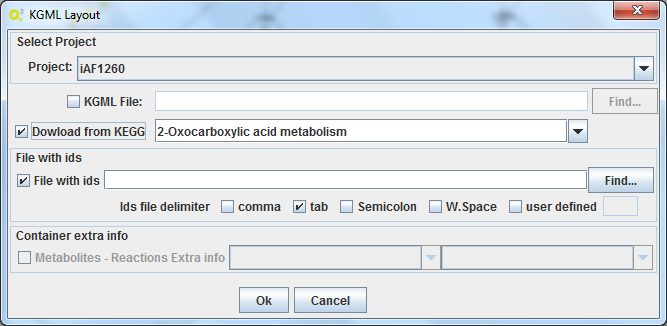
By selecting the Import KGML layout, it is possible to load a KGML file or download it directly from the KEGG database.
This interface also provides the tools to map the metabolic model and the layout. It is possible to submit a 2-column file providing the mapping between the metabolic identifiers and the KEGG ids. If the model loaded in OptFlux has that information, then it is possible to select that option, and the mapping will be made automatically.
File Definitions[edit]
CellDesigner SBML: CellDesigner (http://www.celldesigner.org/) is a well-known tool for drawing cellular networks, based on the process diagram paradigm, with a graphical notation system proposed by Kitano. These are stored using an extension of the Systems Biology Markup Language (SBML).
COBRA Maps: maps developed for COBRA Toolbox. There are, at the moment,several maps on this format for many of the models hosted in the BiGG knowledgebase (http://bigg.ucsd.edu). These maps can be used on different models that have similar pathways, with a correct mapping of the identifiers between the layout and the BiGG model. The files are text tab-delimited files divided in four sections, and since they were created to map a metabolic model for a ME tool, the conversion to the metabolic layout presented in this work was relatively straight forward. These files contain the notion of molecules (metabolites/currency), reaction nodes (transformations) and reactions (reaction nodes).
SBGN PD: SBGN (http://www.sbgn.org/) is a visual language developed by biochemists, modellers and computer scientists that aims at being the standard for visual representation of biological processes.
SBGN defines three visual languages: Process Description (PD), Entity Relationship (ER) and Activity Flow (AF). This plugin supports PD language files. You can learn more about these maps by downloading this file.
At the moment SBGN-PD maps are only supported entities of the type "simple chemical" (for metabolites) and "process" (for reactions). Clone nodes are considered currency metabolites.
You can also learn more about libSGBN here.
KGML: KGML is an exchange format created for the Kyoto encyclopaedia of genes and genomes (KEGG) maps automatic drawing.This format exclusively uses KEGG-ids while genome scale metabolic models do not use these identifiers as primary source of identification of their compounds and reactions, so it is necessary to map the identifiers to make possible the connection between the model and the layout.
This can be made using the Import KGML interface that the plugin provides:
XGMML: This format is based on the Graph Modelling Language (GML), being used for graph description using XML tags to describe nodes and edges of a graph. It is supported, for instance, by Cytoscape (http://www.cytoscape.org/).
To create a metabolic layout from an XGMML file it is necessary to specify some fields for the nodes: the metabolic identifier, node type (metabolite, currency, information, reaction) and, in the case of reaction nodes, the reversibility of that reaction. The exportation of a layout in this format also adds Cytoscape fields, so it is possible to export these generated networks to Cytoscape with a specific visual aspect.
OptFlux xgmml file components should look like this: