(→File Definitions) |
(→File Definitions) |
||
| Line 40: | Line 40: | ||
<br><br> | <br><br> | ||
| − | <b>SBGN PD</b>: | + | <b>SBGN PD</b>: SBGN (http://www.sbgn.org/) is a visual language developed by biochemists, modellers and computer scientists that aims at being the standard for visual representation of biological processes. |
| + | SBGN de�nes three visual languages: Process Description (PD), Entity Relationship (ER) and Activity Flow (AF). This plugin supports PD language �files. You can learn more about these maps by downloading the file: http://sourceforge.net/projects/sbgn/files/SBGN%20Process%20Description%20language/Level%201%20Version%201.3/sbgn_PD-level1-Manual-v1.3.pdf/download. | ||
| + | |||
| + | <br> | ||
| + | |||
| + | At the moment SBGN-PD maps are only supported entities of the type "simple chemical" (for metabolites) and "process" (for reactions). Clone nodes are considered currency metabolites. | ||
<br><br> | <br><br> | ||
Revision as of 14:59, 11 November 2013
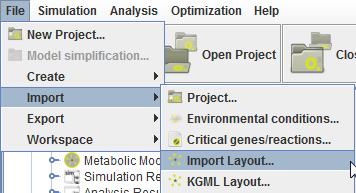
These import layout operations are availabe in the Import menu:
There are 2 major operations for layout importation in OptFlux. The default import operation allows importing layouts from XGMML, COBRA, CellDesigner and SBGN layouts.
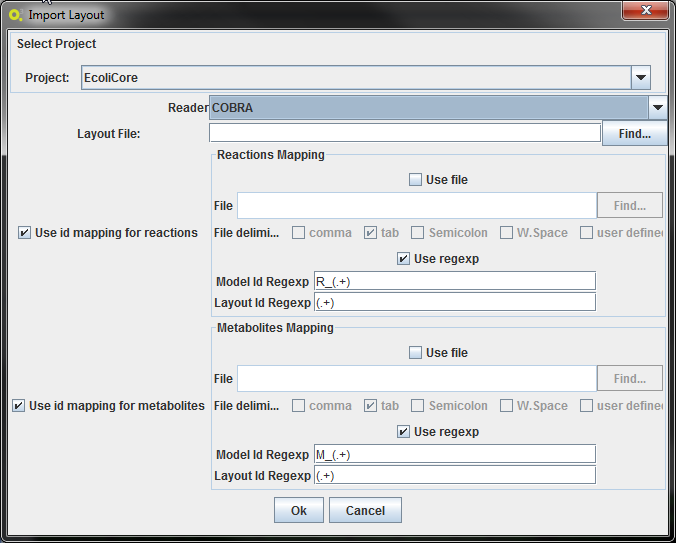
In this interface it is possible to define the mapping of the id's from the metabolic model to the layout. This mapping may not be necessary, but this way it is possible, for instance, to use the same layout for different models.
The mapping can be made two ways:
- Providing a 2-column file: mapping the metabolic model identifier to the layout identifier
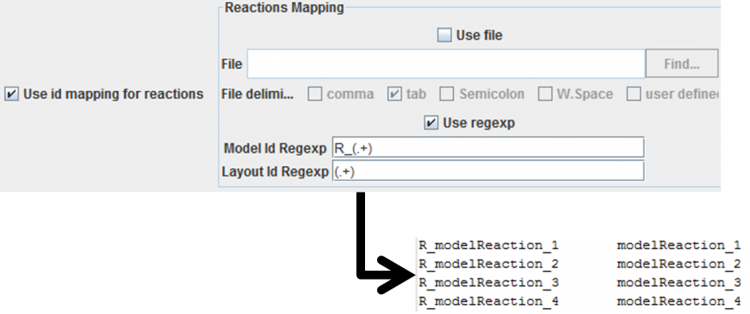
- Applying regexps to the ids: the regexps must contain only one group. If the id's match, by applying those regexp's, then that layout entity will be mapped with the metabolic model entity. An example of a reaction mapping can be seen below.
File Definitions
CellDesigner SBML: CellDesigner (http://www.celldesigner.org/) is a well-known tool for drawing cellular networks, based on the process diagram paradigm, with a graphical notation system proposed by Kitano. These are stored using an extension of the Systems Biology Markup Language (SBML).
COBRA Maps:
SBGN PD: SBGN (http://www.sbgn.org/) is a visual language developed by biochemists, modellers and computer scientists that aims at being the standard for visual representation of biological processes.
SBGN de�nes three visual languages: Process Description (PD), Entity Relationship (ER) and Activity Flow (AF). This plugin supports PD language �files. You can learn more about these maps by downloading the file: http://sourceforge.net/projects/sbgn/files/SBGN%20Process%20Description%20language/Level%201%20Version%201.3/sbgn_PD-level1-Manual-v1.3.pdf/download.
At the moment SBGN-PD maps are only supported entities of the type "simple chemical" (for metabolites) and "process" (for reactions). Clone nodes are considered currency metabolites.
KGML:
XGMML: