| Line 85: | Line 85: | ||
[[Image:RegulatoryNetworkSimulation2.png|800px]] | [[Image:RegulatoryNetworkSimulation2.png|800px]] | ||
| + | |||
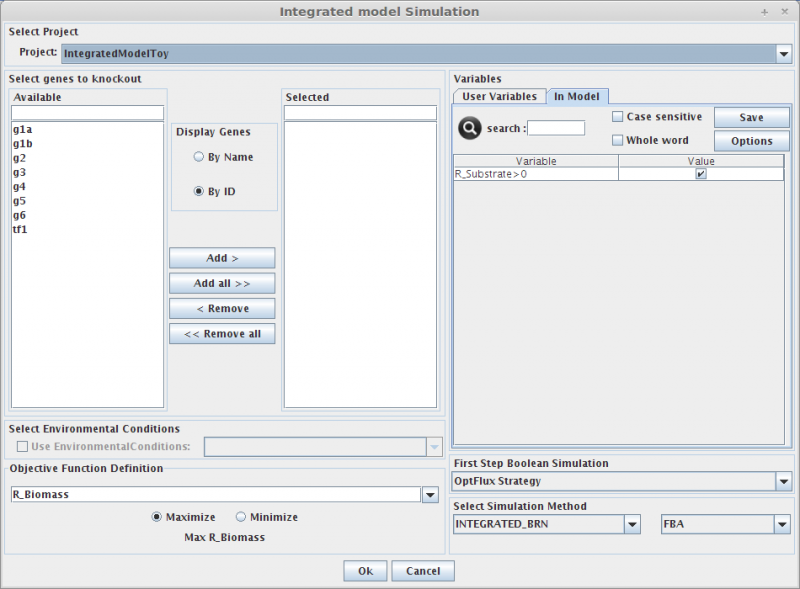
| + | <b>1. <i>Select genes to knockout</i></b><br> | ||
| + | Selecting in the Gene list you can add/remove (using the arrows buttons) genes to the knockout list (the list of genes to be knocked out, in the right).<br> | ||
| + | Genes can be displayed by '''Name''' or '''ID'''. | ||
| + | <br> | ||
| + | <b>2. <i>Select Environmental Conditions</i></b><br> | ||
| + | If you have created [[Environmental conditions|environmental conditions]] you can select them to be used as constraints in the simulation. These can be used to define the values of drain fluxes, i.e. the rates at which metabolites are consumed or produced.<br> | ||
| + | <br> | ||
| + | <b>3. <i>Objective Function Definition </i></b><br> | ||
| + | Here you can select the reaction to optimize (biomass, by default), and you can also define if you will be maximizing or minimizing that flux.<br> | ||
| + | <br> | ||
| + | <b>4. <i>Set Regulatory Variables</i></b><br> | ||
| + | Here you can define the value of all regulatory user variables, and see the values of the model variables.<br> | ||
| + | <br> | ||
| + | <b>5. <i>Select Simulation Method</i></b><br> | ||
| + | OptFlux can use several simulation methods for knockout simulations, namely:<br> | ||
| + | BRN+FBA, BRN+ROOM, BRN+MOMA and SR-FBA<br> | ||
| + | <br> | ||
| + | *And that's all!! You can press OK and the results will be loaded into the clipboard.<br> | ||
| + | |||
=== Integrated Model Simulation === | === Integrated Model Simulation === | ||
[[Image:IntegratedmodelSimulation2.png|800px]] | [[Image:IntegratedmodelSimulation2.png|800px]] | ||
Revision as of 17:12, 12 September 2013
Contents
Example file
You can download it here -> IntegratedModelToy.zip
This tutorial uses an example based on work presented by J. Kim and J. Reed.OptORF: Optimal metabolic and regulatory perturbations for metabolic engineering of microbial strains. BMC, 2010
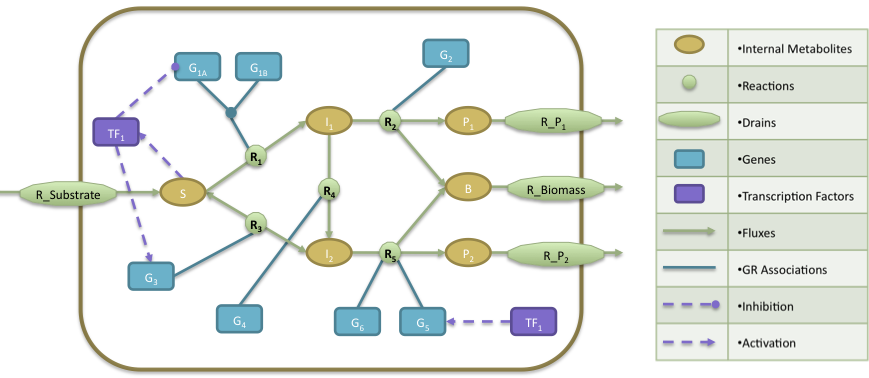
- The regulatory model is composed by a transcription factor (TF1) that is activated when metabolite S is present, activates the expression of two genes (G3, G5) and represses the expression of gene G1A.
- The Gene Reactions rules are characterized in the following way: R1 is catalyzed when the gene G1A and G1B are activated; R2 is catalyzed by the gene G2; R3 and R4 are catalyzed by the genes G3 and G4, respectively; finally, R5 can be catalyzed by either genes G5 or G6.
- At the metabolic level, the substrate (S) is utilized to produce biomass (B) and by-products P1 and P2. The cellular objective is to maximize biomass production (B) and the engineering objective is the production of P1. Reaction R2 converts the internal metabolite I1 into product P1 and 0.08 biomass (B), whereas reaction R5 converts the internal metabolite I2 into product P2 and 0.12 Biomass. The stoichiometric coefficients of all other reactions reflect a one-to-one relationship between molecule quantities.
Build a new project containing an integrated model
The user can build a project with an integrated model in two distinctive ways:
1. Through the "New Project Wizard Operation"
At the moment it is only possible to build an integrated model using the wizard operation from Flat Files, (new Formats will be available in a near future version).
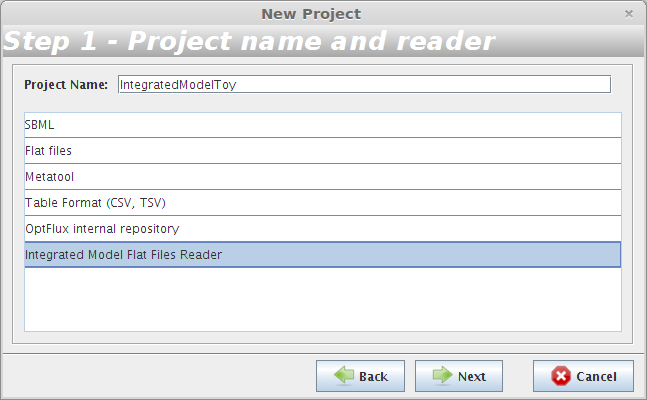
- Step 1
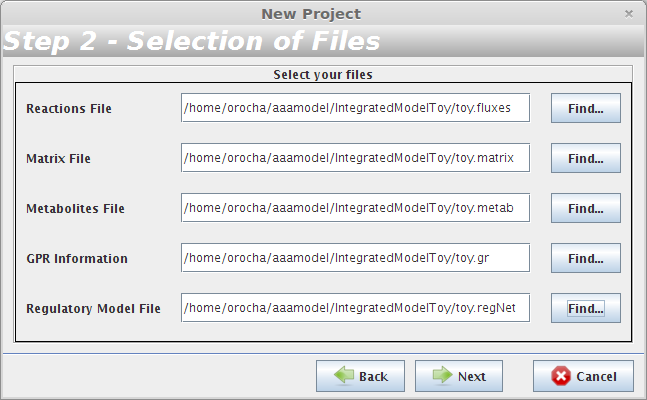
- Step 2
The user have to choose the the toy.fluxes file as the Reactions File; the toy.matrix file as the Matrix File; the toy.metab file as the Metabolites File; the toy.gr file as the GPR Information and the toy.regNet file as the Regulatory Model File.
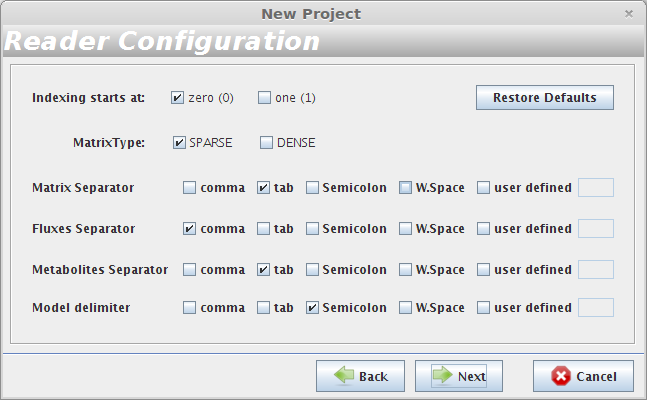
- Reader Configuration
In this step some definitions can be selected (depending from the type of the delimiters that are used on user Flat Files)
Continue to next panel Step 3 by pressing next. The user can select any of the options presented in the panel Step 3 or continue to next panel Step 4.
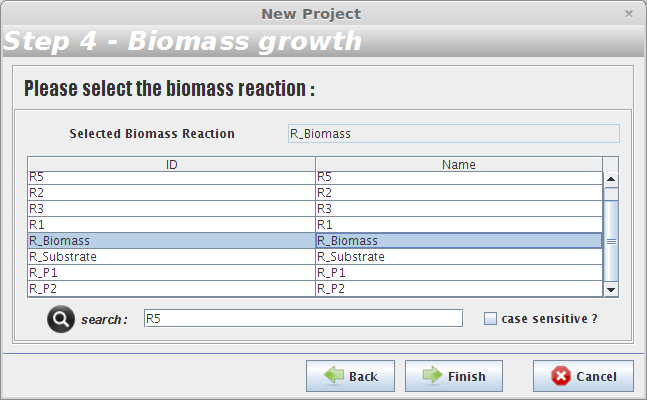
- Step 4
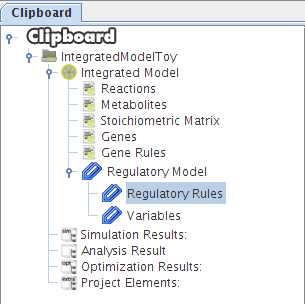
After selecting the biomass reaction, press the Finish button and a new project will be created. The model components will be displayed on Clipboard panel at left.
2. From an existing project
To create an integrated model project by this way it's required to have a metabolic model project already loaded (for more information click here). It's mandatory that the previous loaded metabolic model, have the GPR Rules information.
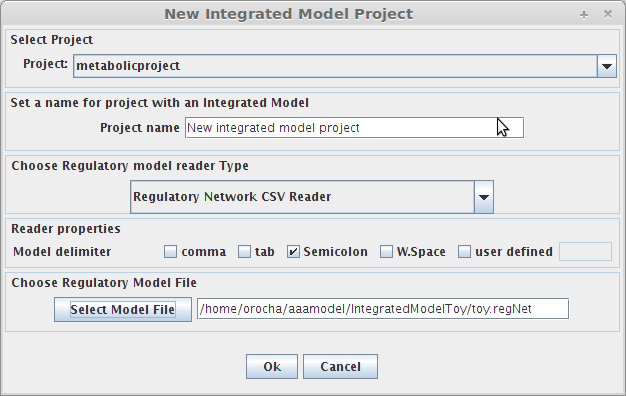
- The following dialog panel will be displayed
1. Project
You can select the model/project to integrate the regulatory model.
2. Choose Regulatory model reader type
You can select the type of reader for the regulatory network.
3. Choose Regulatory model file
You can select the regulatory model file. In our example the regulatory model file name is toy.regNet.
After pressing the Ok button the new project will be created, and the model components will be displayed on Clipboard.
How to perform a mutant simulation
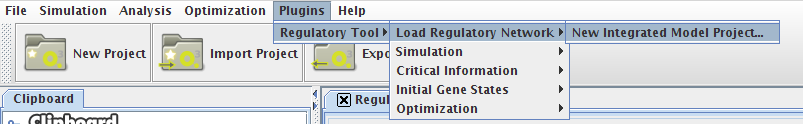
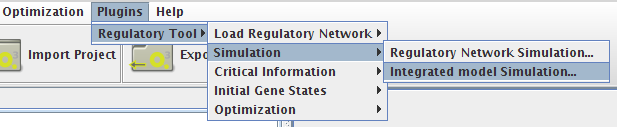
- You can access the Simulation option under the Plugins->Regulatory Tool->Simulation menu or right clicking on the Integrated Model icon on the clipboard.
- In both simulation types you can select the project to work, and set up your configuration.
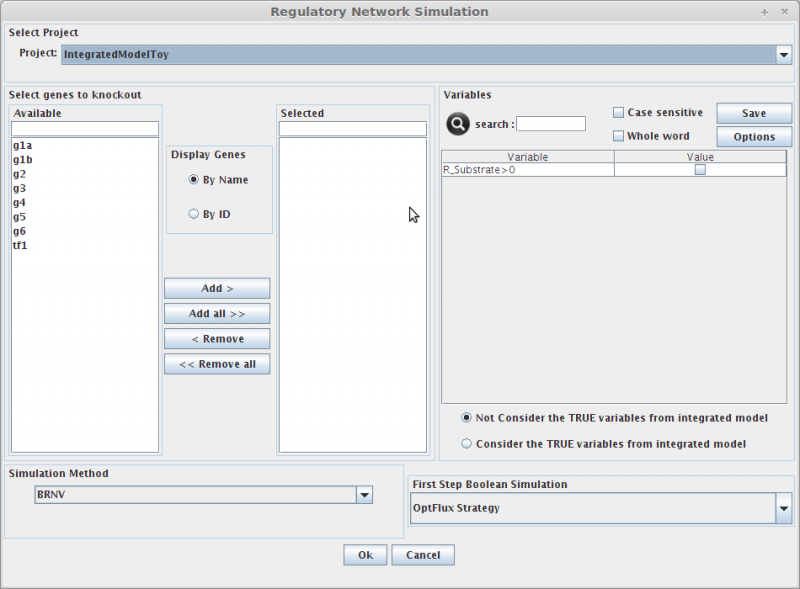
Regulatory Model Simulation
1. Select genes to knockout
Selecting in the Gene list you can add/remove (using the arrows buttons) genes to the knockout list (the list of genes to be knocked out, in the right).
Genes can be displayed by Name or ID.
2. Select Environmental Conditions
If you have created environmental conditions you can select them to be used as constraints in the simulation. These can be used to define the values of drain fluxes, i.e. the rates at which metabolites are consumed or produced.
3. Objective Function Definition
Here you can select the reaction to optimize (biomass, by default), and you can also define if you will be maximizing or minimizing that flux.
4. Set Regulatory Variables
Here you can define the value of all regulatory user variables, and see the values of the model variables.
5. Select Simulation Method
OptFlux can use several simulation methods for knockout simulations, namely:
BRN+FBA, BRN+ROOM, BRN+MOMA and SR-FBA
- And that's all!! You can press OK and the results will be loaded into the clipboard.